| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,399,534 – 16,399,633 |
| Length | 99 |
| Max. P | 0.905629 |

| Location | 16,399,534 – 16,399,633 |
|---|---|
| Length | 99 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.11 |
| Shannon entropy | 0.25233 |
| G+C content | 0.46494 |
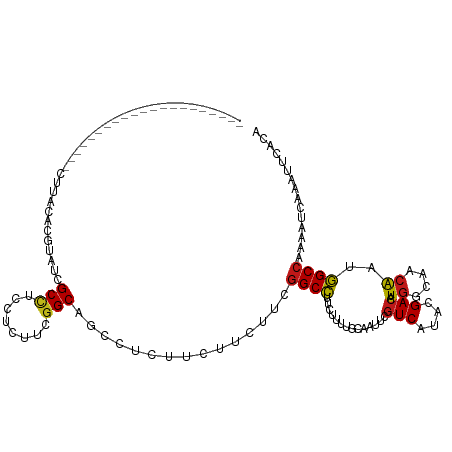
| Mean single sequence MFE | -19.96 |
| Consensus MFE | -15.49 |
| Energy contribution | -15.49 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.905629 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
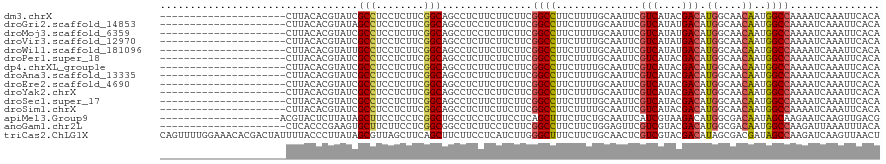
>dm3.chrX 16399534 99 + 22422827 ---------------------CUUACACGUAUCGCCUCCUCUUCGGCAGCCUCUUCUUCUUCGGCCUUCUUUUGCAAUUCGUCAUACGACAUGGCAACAAUGGCCAAAAUCAAAUUCACA ---------------------.......((...(((........)))...............((((.....((((...(((.....)))....))))....))))............)). ( -18.90, z-score = -2.39, R) >droGri2.scaffold_14853 3252910 99 - 10151454 ---------------------CUUACACGUAUAGCCUCCUCUUCGGCAGCCUCCUCUUCUUCGGCCUUCUUUUGCAAUUCGUCAUAUGACAUGGCAACAAUGGCCAAAAUCAAAUUCACA ---------------------.......((...(((........)))...............((((.....((((.....(((....)))...))))....))))............)). ( -18.60, z-score = -1.85, R) >droMoj3.scaffold_6359 3319143 99 + 4525533 ---------------------CUUACACGUAUCGCCUCCUCUUCGGCAGCCUCCUCUUCUUCGGCCUUCUUUUGCAAUUCGUCAUAUGACAUGGCAACAAUGGCCAAAAUCAAAUUCACA ---------------------.......((...(((........)))...............((((.....((((.....(((....)))...))))....))))............)). ( -18.70, z-score = -2.05, R) >droVir3.scaffold_12970 2104524 99 + 11907090 ---------------------CUUACACGUAUCGCCUCCUCUUCGGCAGCCUCUUCUUCUUCGGCCUUCUUUUGCAAUUCGUCAUAUGACAUGGCAACAAUGGCCAAAAUCAAAUUCACA ---------------------.......((...(((........)))...............((((.....((((.....(((....)))...))))....))))............)). ( -18.70, z-score = -2.17, R) >droWil1.scaffold_181096 5817314 99 + 12416693 ---------------------CUUACACGUAUUGCCUCCUCUUCGGCAGCCUCUUCUUCUUCGGCCUUCUUUUGCAAUUCGUCAUAUGACAUGGCAACAAUGGCCAAAAUCAAAUUCACA ---------------------.......((.(((((........))))).............((((.....((((.....(((....)))...))))....))))............)). ( -19.70, z-score = -2.33, R) >droPer1.super_18 15699 99 - 1952607 ---------------------CUUACACGUAUCGCCUCCUCUUCGGCAGCCUCUUCUUCUUCGGCCUUCUUUUGCAAUUCGUCAUACGACAUGGCAACAAUGGCCAAAAUCAAAUUCACA ---------------------.......((...(((........)))...............((((.....((((...(((.....)))....))))....))))............)). ( -18.90, z-score = -2.39, R) >dp4.chrXL_group1e 9496776 99 - 12523060 ---------------------CUUACACGUAUCGCCUCCUCUUCGGCAGCCUCUUCUUCUUCGGCCUUCUUUUGCAAUUCGUCAUACGACAUGGCAACAAUGGCCAAAAUCAAAUUCACA ---------------------.......((...(((........)))...............((((.....((((...(((.....)))....))))....))))............)). ( -18.90, z-score = -2.39, R) >droAna3.scaffold_13335 1791638 99 - 3335858 ---------------------CUUACACGUAUCGCCUCCUCUUCGGCAGCCUCUUCUUCUUCGGCCUUCUUUUGCAAUUCGUCAUACGACAUGGCAACAAUGGCCAAAAUCAAAUUCACA ---------------------.......((...(((........)))...............((((.....((((...(((.....)))....))))....))))............)). ( -18.90, z-score = -2.39, R) >droEre2.scaffold_4690 8044456 99 - 18748788 ---------------------CUUACACGUAUCGCCUCCUCUUCGGCAGCCUCUUCUUCUUCGGCCUUCUUUUGCAAUUCGUCAUACGACAUGGCAACAAUGGCCAAAAUCAAAUUCACA ---------------------.......((...(((........)))...............((((.....((((...(((.....)))....))))....))))............)). ( -18.90, z-score = -2.39, R) >droYak2.chrX 10514789 99 + 21770863 ---------------------CUUACACGUAUCGCCUCCUCUUCGGCAGCCUCCUCUUCUUCGGCCUUCUUUUGCAAUUCGUCAUACGACAUGGCAACAAUGGCCAAAAUCAAAUUCACA ---------------------.......((...(((........)))...............((((.....((((...(((.....)))....))))....))))............)). ( -18.90, z-score = -2.29, R) >droSec1.super_17 213237 99 + 1527944 ---------------------CUUACACGUAUCGCCUCCUCUUCGGCAGCCUCUUCUUCUUCGGCCUUCUUUUGCAAUUCGUCAUACGACAUGGCAACAAUGGCCAAAAUCAAAUUCACA ---------------------.......((...(((........)))...............((((.....((((...(((.....)))....))))....))))............)). ( -18.90, z-score = -2.39, R) >droSim1.chrX 12598523 99 + 17042790 ---------------------CUUACACGUAUCGCCUCCUCUUCGGCAGCCUCUUCUUCUUCGGCCUUCUUUUGCAAUUCGUCAUACGACAUGGCAACAAUGGCCAAAAUCAAAUUCACA ---------------------.......((...(((........)))...............((((.....((((...(((.....)))....))))....))))............)). ( -18.90, z-score = -2.39, R) >apiMel3.Group9 4303995 100 - 10282195 --------------------ACGUACUCUUAUAGCUUCCUCCUCGGCUGCCUCCUCUUCCUCAGCUUUCUUCUGCAAUUCAUCGUAAGACAUGGCGACAAUAGCAAGAAUCAAGUUGACG --------------------...........(((((........)))))...........(((((((..((((((......((....))..((....))...)).))))..))))))).. ( -18.30, z-score = -0.46, R) >anoGam1.chr2L 46921859 99 - 48795086 ---------------------CUCACCCGAAGUGCUUCUUCCUCGGCGGCCUCUUCCUCUUCGGCCUUCUUCUGGAGUUCGUCGUACGACAUGGCGACAAUGGCCAAGAUUAAAUUUACA ---------------------.....((((.(........).)))).((((...........(..((((....))))..)(((((........)))))...))))............... ( -23.50, z-score = 0.02, R) >triCas2.ChLG1X 5770128 120 - 8109244 CAGUUUUGGAAACACGACUAUUUUACCCUUAUAGCGUUAGCUUCAGCUUCUUCCUCAUCUUGGGCUUUCUUCUGCAACUCGUCGUACGACAUAGCGACGAUAGCCAAGAUCAAGUUAACU .((((.((....)).))))................((((((((.............(((((((((........))...(((((((........)))))))...))))))).)))))))). ( -30.74, z-score = -2.56, R) >consensus _____________________CUUACACGUAUCGCCUCCUCUUCGGCAGCCUCUUCUUCUUCGGCCUUCUUUUGCAAUUCGUCAUACGACAUGGCAACAAUGGCCAAAAUCAAAUUCACA .................................(((........)))...............((((..............(((....))).((....))..))))............... (-15.49 = -15.49 + 0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:49:45 2011