| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,399,226 – 16,399,351 |
| Length | 125 |
| Max. P | 0.997402 |

| Location | 16,399,226 – 16,399,351 |
|---|---|
| Length | 125 |
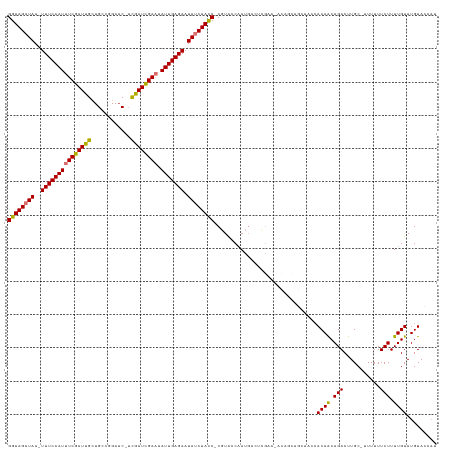
| Sequences | 8 |
| Columns | 129 |
| Reading direction | reverse |
| Mean pairwise identity | 83.60 |
| Shannon entropy | 0.30467 |
| G+C content | 0.37272 |
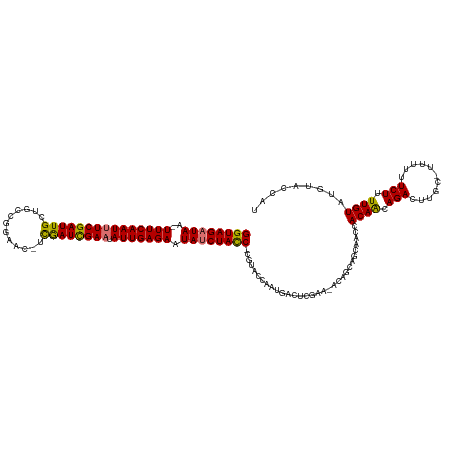
| Mean single sequence MFE | -29.16 |
| Consensus MFE | -21.28 |
| Energy contribution | -21.07 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.30 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.09 |
| SVM RNA-class probability | 0.997402 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
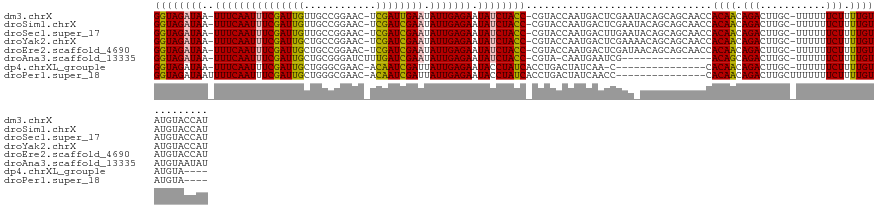
>dm3.chrX 16399226 125 - 22422827 GGUAGAUAA-UUUCAAUUUCGAUUGUUGCCGGAAC-UCGAUUGAAUAUUGAGAAUAUCUACC-CGUACCAAUGACUCGAAUACAGCAGCAACCACAACAGACUUGC-UUUUUUCUUUUGUAUGUACCAU ((((((((.-(((((((((((((((..........-.)))))))).))))))).))))))))-.((((..(..(...(((...(((((..............))))-)...)))..)..)..))))... ( -28.74, z-score = -2.77, R) >droSim1.chrX 12598218 125 - 17042790 GGUAGAUAA-UUUCAAUUUCGAUUGUUGCCGGAAC-UCGAUCGAAUAUUGAGAAUAUCUACC-CGUACCAAUGACUCGAAUACAGCAGCAACCACAACAGACUUGC-UUUUUUCUUUUGUAUGUACCAU ((((((((.-(((((((((((((((..........-.)))))))).))))))).))))))))-.((((..(..(...(((...(((((..............))))-)...)))..)..)..))))... ( -30.84, z-score = -3.37, R) >droSec1.super_17 212932 125 - 1527944 GGUAGAUAA-UUUCAAUUUCGAUUGUUGCCGGAAC-UCGAUCGAAUAUUGAGAAUAUCUACC-CGUACCAAUGACUUGAAUACAGCAGCAACCACAACAGACUUGC-UUUUUUCUUUUGUAUGUACCAU ((((((((.-(((((((((((((((..........-.)))))))).))))))).))))))))-.((((..(..(...(((...(((((..............))))-)...)))..)..)..))))... ( -31.54, z-score = -3.69, R) >droYak2.chrX 10514466 125 - 21770863 GGUAGAUAA-UUUCAAUUUCGAUUGCUGCCGGAAC-UCGAUCGAAUAUUGAGAAUAUCUACC-CGUACCAAUGACUCGAAAACAGCAGCAACCACAACAGACUUGC-UUUUUUCUUUUGUAUGUACCAU ((((((((.-(((((((((((((((..(......)-.)))))))).))))))).))))))))-.((((..(..(...(((((.(((((..............))))-).)))))..)..)..))))... ( -32.94, z-score = -4.15, R) >droEre2.scaffold_4690 8044150 125 + 18748788 GGUAGAUAA-UUUCAAUUUCGAUUGCUGCCGGAAC-UCGAUCGAAUAUUGAGAAUAUCUACC-CGUACCAAUGACUCGAUAACAGCAGCAACCACAACAGACUUGC-UUUUUUCUUUUGUAUGUACCAU ((((((((.-(((((((((((((((..(......)-.)))))))).))))))).))))))))-.((((..(..(...((.((.(((((..............))))-).)).))..)..)..))))... ( -29.64, z-score = -3.06, R) >droAna3.scaffold_13335 1791267 110 + 3335858 GGUAGAUAA-UUUCAAUUUCGAUUGCUGCGGGAUCUUUGAUCGAAUAUUGAGAAUAUCUACC-CGUA-CAAUGAAUCG---------------ACAGCAGACUUGC-UUUUUUCUUUUGUAUGUAAUAU ((((((((.-(((((((((((((((..(......)..)))))))).))))))).))))))))-((((-(((.(((...---------------..(((......))-)...)))..)))))))...... ( -32.10, z-score = -3.86, R) >dp4.chrXL_group1e 9496419 106 + 12523060 GGUAGAUAA-UUUCAAUUUCGAUUGCUGGGCGAAC-ACAAUCGAUUAUUGAGAAUACCUAUCACCUGACUAUCAA-C---------------CACAACAGACUUGC-UUUUUUCUUUUGUAUGUA---- (((((.((.-(((((((.(((((((.((......)-))))))))..))))))).)).))))).............-.---------------.((((.(((.....-.....))).)))).....---- ( -23.20, z-score = -2.56, R) >droPer1.super_18 15337 109 + 1952607 GGUAGAUAAUUUUCAAUUUCGAUUGCUGGGCGAAC-ACAAUCGAUUAUUGAGAAUACCUAUCACCUGACUAUCAACC---------------CACAACAGACUUGCUUUUUUUCUUUUGUAUGUA---- (((.(((((((((((((.(((((((.((......)-))))))))..)))))))))...))))))).........((.---------------.((((.(((...........))).))))..)).---- ( -24.30, z-score = -2.93, R) >consensus GGUAGAUAA_UUUCAAUUUCGAUUGCUGCCGGAAC_UCGAUCGAAUAUUGAGAAUAUCUACC_CGUACCAAUGACUCGAA_ACAGCAGCAACCACAACAGACUUGC_UUUUUUCUUUUGUAUGUACCAU (((((((...(((((((((((((((............)))))))).)))))))..)))))))...............................((((.(((...........))).))))......... (-21.28 = -21.07 + -0.20)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:49:44 2011