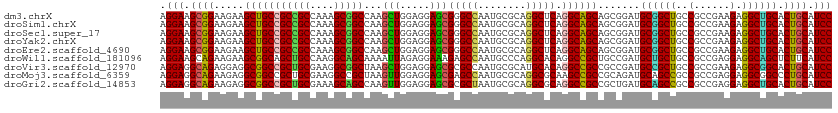
| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,398,221 – 16,398,333 |
| Length | 112 |
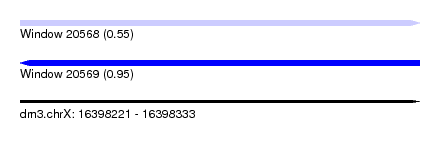
| Max. P | 0.950940 |

| Location | 16,398,221 – 16,398,333 |
|---|---|
| Length | 112 |
| Sequences | 9 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 87.95 |
| Shannon entropy | 0.24975 |
| G+C content | 0.68651 |
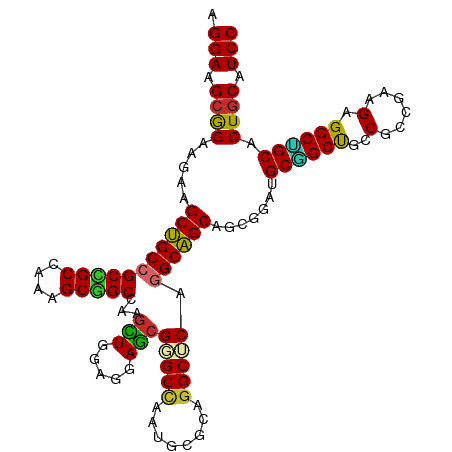
| Mean single sequence MFE | -56.35 |
| Consensus MFE | -38.37 |
| Energy contribution | -38.36 |
| Covariance contribution | -0.01 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.553574 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
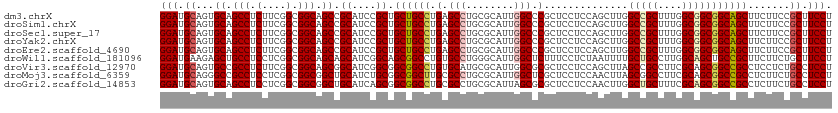
>dm3.chrX 16398221 112 + 22422827 GGAUGCAGUGCAGCCUCUUCGGCGGCAGCCGCAUCCGCUGCUGCCUGAGCCUGCGCAUUGGCCCGCUCCUCCAGCUUGGCCGCUUUGGCGGCGGCAGCUUCUUCCGCUUCCU (((.((.((((((.(((...(((((((((.......))))))))).))).))))))....(((.(((.....)))...(((((....))))))))..........)).))). ( -55.10, z-score = -1.41, R) >droSim1.chrX 12597228 112 + 17042790 GGAUGCAGUGCAGCCUCUUCGGCGGCAGCCGCAUCCGCUGCUGCCUGAGCCUGCGCAUUGGCCCGCUCCUCCAGCUUGGCCGCUUUGGCGGCGGCAGCUUCUUCCGCUUCCU (((.((.((((((.(((...(((((((((.......))))))))).))).))))))....(((.(((.....)))...(((((....))))))))..........)).))). ( -55.10, z-score = -1.41, R) >droSec1.super_17 211969 112 + 1527944 GGAUGCAGUGCAGCCUCUUCGGCGGCAGCCGCAUCCGCUGCUGCCUGAGCCUGCGCAUUGGCCCGCUCCUCCAGCUUGGCCGCUUUGGCGGCGGCAGCUUCUUCCGCUUCCU (((.((.((((((.(((...(((((((((.......))))))))).))).))))))....(((.(((.....)))...(((((....))))))))..........)).))). ( -55.10, z-score = -1.41, R) >droYak2.chrX 10513461 112 + 21770863 GGAUGCAGUGCAGCCUCUUCGGCGGCAGCCGCAUCCGCUGCUGCCUGAGCCUGCGCAUUGGCCCGCUCCUCCAGCUUGGCCGCUUUGGCGGCGGCAGCUUCUUCCGCUUCCU (((.((.((((((.(((...(((((((((.......))))))))).))).))))))....(((.(((.....)))...(((((....))))))))..........)).))). ( -55.10, z-score = -1.41, R) >droEre2.scaffold_4690 8043207 112 - 18748788 GGAUGCAGUGCAGCCUCUUCGGCGGCAGCCGCAUCCGCUGCUGCCUGAGCCUGCGCAUUGGCCCGCUCCUCCAGCUUGGCCGCUUUGGCGGCGGCAGCUUCUUCCGCUUCCU (((.((.((((((.(((...(((((((((.......))))))))).))).))))))....(((.(((.....)))...(((((....))))))))..........)).))). ( -55.10, z-score = -1.41, R) >droWil1.scaffold_181096 5814485 112 + 12416693 GGAUGAAGAGCUGCCUCCUCGGCGGCAGCAGCAUCGGCAGCGGCCUGUGCCUGGGCAUUGGCUCUUUCCUCUAAUUUUGCUGCCUUGGCAGCUGCCGCUUCUUCUGCUUCCU (((.(((((((.(((.....)))((((((.((...((((((((((.((((....)))).))))...............))))))...)).)))))))).)))))....))). ( -52.96, z-score = -2.54, R) >droVir3.scaffold_12970 2103187 112 + 11907090 GGAUGCAGUGCCGCCUCUUCGGCGGCAGCGGCAUCGGCGGCGGCCUGUGCAUGCGCAUUGGCGCGCUCCUCCAGCUUAGCCGCCUUCGCAGCGGCCGCCUCCUCUGCCUCCU (((.((..(((((((.....)))))))))((((..((.(((((((...((.((((....((((.(((..........)))))))..))))))))))))).))..))))))). ( -60.60, z-score = -2.15, R) >droMoj3.scaffold_6359 3317840 112 + 4525533 GGAUGCAGGGCCGCCUCCUCGGCGGCGGCUGCAUCUGCGGCGGCUUGCGCCUGCGCAUUGGCUCGCUCCUCCAACUUAGCGGCCUUCGCAGCGGCCGCCUCUUCUGCCUCCU (((.(((((((((......))))(((((((((....((((((.....)))).))((...(((.((((..........)))))))...)).)))))))))...))))).))). ( -58.50, z-score = -2.23, R) >droGri2.scaffold_14853 3251558 112 - 10151454 GGAUGCAGUGCAGCCUCCUCGGCGGCGGCUGCAUCAGCGGCGGCCUGCGCCUGCGCAUUAGCGCGCUCCUCCAACUUGGCUGCUUUCGCAGCGGCCGCCUCUUCUGCCUCCU (((.(((((((((((.((.....)).)))))))..((.(((((((.((((..((......))))))............(((((....)))))))))))).)).)))).))). ( -59.60, z-score = -2.77, R) >consensus GGAUGCAGUGCAGCCUCUUCGGCGGCAGCCGCAUCCGCUGCUGCCUGAGCCUGCGCAUUGGCCCGCUCCUCCAGCUUGGCCGCUUUGGCGGCGGCAGCUUCUUCCGCUUCCU .(((((.((((.(((.....))).))).).)))))....((((((.(.(((........))).)..............(((((....))))))))))).............. (-38.37 = -38.36 + -0.01)
| Location | 16,398,221 – 16,398,333 |
|---|---|
| Length | 112 |
| Sequences | 9 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 87.95 |
| Shannon entropy | 0.24975 |
| G+C content | 0.68651 |
| Mean single sequence MFE | -58.52 |
| Consensus MFE | -48.02 |
| Energy contribution | -46.98 |
| Covariance contribution | -1.05 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.950940 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
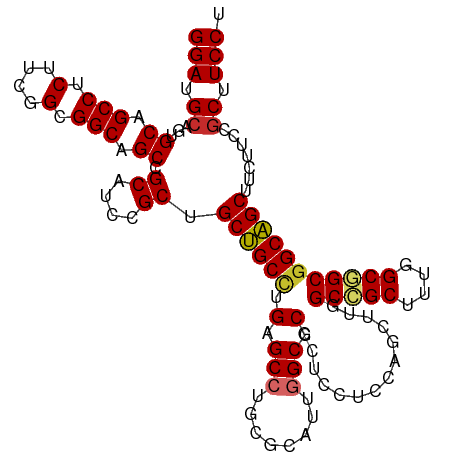
>dm3.chrX 16398221 112 - 22422827 AGGAAGCGGAAGAAGCUGCCGCCGCCAAAGCGGCCAAGCUGGAGGAGCGGGCCAAUGCGCAGGCUCAGGCAGCAGCGGAUGCGGCUGCCGCCGAAGAGGCUGCACUGCAUCC ......(....)..(((((((((((....)))))...(((.....)))(((((........))))).))))))...((((((((.(((.(((.....))).))))))))))) ( -56.60, z-score = -1.29, R) >droSim1.chrX 12597228 112 - 17042790 AGGAAGCGGAAGAAGCUGCCGCCGCCAAAGCGGCCAAGCUGGAGGAGCGGGCCAAUGCGCAGGCUCAGGCAGCAGCGGAUGCGGCUGCCGCCGAAGAGGCUGCACUGCAUCC ......(....)..(((((((((((....)))))...(((.....)))(((((........))))).))))))...((((((((.(((.(((.....))).))))))))))) ( -56.60, z-score = -1.29, R) >droSec1.super_17 211969 112 - 1527944 AGGAAGCGGAAGAAGCUGCCGCCGCCAAAGCGGCCAAGCUGGAGGAGCGGGCCAAUGCGCAGGCUCAGGCAGCAGCGGAUGCGGCUGCCGCCGAAGAGGCUGCACUGCAUCC ......(....)..(((((((((((....)))))...(((.....)))(((((........))))).))))))...((((((((.(((.(((.....))).))))))))))) ( -56.60, z-score = -1.29, R) >droYak2.chrX 10513461 112 - 21770863 AGGAAGCGGAAGAAGCUGCCGCCGCCAAAGCGGCCAAGCUGGAGGAGCGGGCCAAUGCGCAGGCUCAGGCAGCAGCGGAUGCGGCUGCCGCCGAAGAGGCUGCACUGCAUCC ......(....)..(((((((((((....)))))...(((.....)))(((((........))))).))))))...((((((((.(((.(((.....))).))))))))))) ( -56.60, z-score = -1.29, R) >droEre2.scaffold_4690 8043207 112 + 18748788 AGGAAGCGGAAGAAGCUGCCGCCGCCAAAGCGGCCAAGCUGGAGGAGCGGGCCAAUGCGCAGGCUCAGGCAGCAGCGGAUGCGGCUGCCGCCGAAGAGGCUGCACUGCAUCC ......(....)..(((((((((((....)))))...(((.....)))(((((........))))).))))))...((((((((.(((.(((.....))).))))))))))) ( -56.60, z-score = -1.29, R) >droWil1.scaffold_181096 5814485 112 - 12416693 AGGAAGCAGAAGAAGCGGCAGCUGCCAAGGCAGCAAAAUUAGAGGAAAGAGCCAAUGCCCAGGCACAGGCCGCUGCCGAUGCUGCUGCCGCCGAGGAGGCAGCUCUUCAUCC .(((..........((((((((.((...((((((....((....))..(.(((..(((....)))..))))))))))...)).)))))))).((((((....)))))).))) ( -50.20, z-score = -2.39, R) >droVir3.scaffold_12970 2103187 112 - 11907090 AGGAGGCAGAGGAGGCGGCCGCUGCGAAGGCGGCUAAGCUGGAGGAGCGCGCCAAUGCGCAUGCACAGGCCGCCGCCGAUGCCGCUGCCGCCGAAGAGGCGGCACUGCAUCC .(((((((..((.((((((((((((....)))))...(((.....)))((((....)))).......))))))).))..))))(((((((((.....)))))))..)).))) ( -68.00, z-score = -3.99, R) >droMoj3.scaffold_6359 3317840 112 - 4525533 AGGAGGCAGAAGAGGCGGCCGCUGCGAAGGCCGCUAAGUUGGAGGAGCGAGCCAAUGCGCAGGCGCAAGCCGCCGCAGAUGCAGCCGCCGCCGAGGAGGCGGCCCUGCAUCC .(((.((((....(((.((..(((((..(((((((..........)))).))).....((.(((....))))))))))..)).)))((((((.....)))))).)))).))) ( -59.60, z-score = -2.34, R) >droGri2.scaffold_14853 3251558 112 + 10151454 AGGAGGCAGAAGAGGCGGCCGCUGCGAAAGCAGCCAAGUUGGAGGAGCGCGCUAAUGCGCAGGCGCAGGCCGCCGCUGAUGCAGCCGCCGCCGAGGAGGCUGCACUGCAUCC .(((.((((.((.((((((((((((....)))(((..(((.....)))((((....)))).))))).))))))).))..(((((((.((.....)).))))))))))).))) ( -65.90, z-score = -3.88, R) >consensus AGGAAGCGGAAGAAGCUGCCGCCGCCAAAGCGGCCAAGCUGGAGGAGCGGGCCAAUGCGCAGGCUCAGGCAGCAGCGGAUGCGGCUGCCGCCGAAGAGGCUGCACUGCAUCC .(((.((((.....(((((((((((....)))))...(((.....)))(((((........))))).)))))).......((((((..(......).)))))).)))).))) (-48.02 = -46.98 + -1.05)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:49:43 2011