
| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,376,737 – 16,376,851 |
| Length | 114 |
| Max. P | 0.500000 |

| Location | 16,376,737 – 16,376,851 |
|---|---|
| Length | 114 |
| Sequences | 14 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 85.60 |
| Shannon entropy | 0.33950 |
| G+C content | 0.47500 |
| Mean single sequence MFE | -32.49 |
| Consensus MFE | -23.73 |
| Energy contribution | -22.86 |
| Covariance contribution | -0.87 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chrX 16376737 114 - 22422827 ---UAUACUCGAUGAAUAUCCAGCUGAUUGCUGCCCCGAUUCGUACUAUAAGAAAUUUCCGAUCUUAGCCGGUGACGAUGACUCGCCGUUCUGGCAAGGAUGGGGCAAUUUACGACU ---.....(((((((((...((((.....)))).....))))))........((((((((.(((((.(((((.((((.........))))))))).))))).))).))))).))).. ( -32.50, z-score = -1.51, R) >droSim1.chrX 12588216 114 - 17042790 ---UAUACUCGAUGAAUAUCCAGCUGAUUGCUGCCCCGAUUCGUACUAUAAGAAAUUUCCGAUCUUAGCCGGUGACGAUGACUCGCCGUUCUGGCAAGGAUGGGGCAAUUUACGACU ---.....(((((((((...((((.....)))).....))))))........((((((((.(((((.(((((.((((.........))))))))).))))).))).))))).))).. ( -32.50, z-score = -1.51, R) >droSec1.super_17 196254 114 - 1527944 ---UAUACUCGAUGAAUAUCCAGCUGAUUGCUGCCCCGAUUCGUACUAUAAGAAAUUUCCGAUCUUAGCCGGUGACGAUGACUCGCCGUUCUGGCAAGGAUGGGGCAAUUUACGACU ---.....(((((((((...((((.....)))).....))))))........((((((((.(((((.(((((.((((.........))))))))).))))).))).))))).))).. ( -32.50, z-score = -1.51, R) >droYak2.chrX 10495715 114 - 21770863 ---UAUACUCGAUGAAUAUCCAGCUGAUUGCUGCCCCGAUUCGUACUAUAAGAAAUUUCCGAUCUUAGCCGGUGACGAUGACUCGCCGUUCUGGCAAGGAUGGGGCAAUUUACGACU ---.....(((((((((...((((.....)))).....))))))........((((((((.(((((.(((((.((((.........))))))))).))))).))).))))).))).. ( -32.50, z-score = -1.51, R) >droEre2.scaffold_4690 8027391 114 + 18748788 ---UAUACUCGAUGAAUAUCCAGCUGAUUGCUGCCCCGAUUCGUACUAUAAGAAAUUUCCGAUCUUAGCCGGUGACGAUGACUCGCCGUUCUGGCAAGGAUGGGGCAAUUUACGACU ---.....(((((((((...((((.....)))).....))))))........((((((((.(((((.(((((.((((.........))))))))).))))).))).))))).))).. ( -32.50, z-score = -1.51, R) >droAna3.scaffold_13335 1773885 114 + 3335858 ---UAUACUCGAUGAAUAUCCAGCUGAUUGCUGCCCCGAUUCGUACUAUAAGAAAUUUCCGAUCUUAGCCGGUGACGAAGACUCGCCGUUCUGGCAAGGAUGGGGCAAUUUACGACU ---.....(((((((((...((((.....)))).....))))))........((((((((.(((((.(((((.((((.........))))))))).))))).))).))))).))).. ( -32.50, z-score = -1.49, R) >dp4.chrXL_group1e 9479135 114 + 12523060 ---UAUACUCGACGAAUAUCCAGCUGAUUGCUGCCCCGAUUCGUACUAUAAGAAAUUUCCGAUCUUAGCCGGUGAUGAUGACUCGCCAUUCUGGCAAGGAUGGGGCAAUUUACGACU ---.....(((((((((...((((.....)))).....))))))........((((((((.(((((.(((((.((((.........))))))))).))))).))).))))).))).. ( -32.90, z-score = -1.94, R) >droWil1.scaffold_181096 5795706 114 - 12416693 ---UAUAUUGGACGAGUAUCCAGCUGAUUGCUGCCCCGAUUCAUACUAUAAGAAAUUUCCAAUCUUAGCCGGUGACGAAGAUUCGCCGUUCUGGCAAGGAUGGGGCAAUUUACGACU ---.....((((......))))((.....))((((((.((((......(((((.........)))))(((((.((((.........)))))))))..)))))))))).......... ( -31.50, z-score = -1.11, R) >droVir3.scaffold_12970 2084061 114 - 11907090 ---UAUACUUGACGAGUAUCCAGCUGAUUGCUGCCCCGAUUCGUACUAUAAGAAAUUUCCGAUCUUAGCCGGUGACGAAGAUUCGCCGUUCUGGCAAGGAUGGGGCAAUUUACGACU ---....((((((((((...((((.....)))).....))))))....))))((((((((.(((((.(((((.((((.........))))))))).))))).))).)))))...... ( -32.80, z-score = -1.40, R) >droMoj3.scaffold_6359 3297959 114 - 4525533 ---UAUACUUGACGAAUAUCCAGCUGAUUGCUGCCCCGAUUCGUACUAUAAGAAAUUUCCAAUCUUAGCCGGUGACGAAGAUUCGCCGUUCUGGCAAGGAUGGGGCAAUUUACGACU ---....((((((((((...((((.....)))).....))))))....))))((((((((.(((((.(((((.((((.........))))))))).))))).))).)))))...... ( -30.70, z-score = -1.30, R) >droGri2.scaffold_14853 3232850 114 + 10151454 ---UAUACUUGACGAGUAUCCAGCUGAUUGCUGCCCCGAUUCGUACUAUAAGAAAUUUCCAAUCUUAGCCGGGGACGAUGAUUCGCCGUUCUGGCAAGGAUGGGGCAAUUUACGACU ---....((((((((((...((((.....)))).....))))))....))))((((((((.(((((.(((((((.(((....)))...))))))).))))).))).)))))...... ( -32.00, z-score = -0.94, R) >anoGam1.chr2L 46888052 114 + 48795086 ---AGUGAUUGAGGAUUCACCGGCGGAUUGCUGCCCGGACAACUGCUACAAAAAAUUUCCUGUUCUUGCUGGGGAUGAUGACGCGCCGUUCUGGCAGGGUUGGGGAAAUUUACGUCU ---(((..(((.((.....))(((((....)))))....)))..))).....(((((((((..((((((..((.(((.........)))))..))))))...)))))))))...... ( -32.30, z-score = 0.46, R) >apiMel3.Group9 4293335 117 + 10282195 UCCCAUCGAAUCCGAUUAUCCAGCCGAUUGUUGUCCAGAAAAUUGUUACAAGAAGUUCCCAUUCUUGGCUGGCGACGACGACGCACCGUUUUGGCAGGGUUGGGCUAAUUUGAGACU .(((((((....)))..((((.(((((..((((.((((..........((((((.......)))))).))))))))((((......))))))))).))))))))............. ( -31.80, z-score = -0.30, R) >triCas2.ChLG1X 5765703 117 + 8109244 UAUAAUUAUCGACGAUUACGCAGCCGACUGCUUUCCCGAUUCAUGUUACAAGAAGUUUCCGUUUCUUGCAGGCGACGAGGAUUCGCCGUUUUGGCAGGGUUGGGGGAAUUUAAGGUC ..........(((......((((....))))(..((((((((.((((((((((((......)))))))..(((((.......)))))....)))))))))))))..).......))) ( -35.80, z-score = -1.42, R) >consensus ___UAUACUCGACGAAUAUCCAGCUGAUUGCUGCCCCGAUUCGUACUAUAAGAAAUUUCCGAUCUUAGCCGGUGACGAUGACUCGCCGUUCUGGCAAGGAUGGGGCAAUUUACGACU ...........((((((...((((.....)))).....))))))........(((((((((.((((.(((((.((((.........))))))))))))).))))..)))))...... (-23.73 = -22.86 + -0.87)
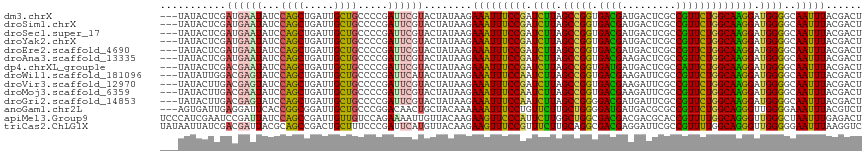
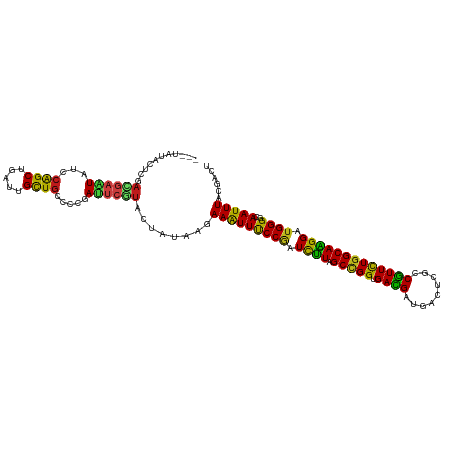
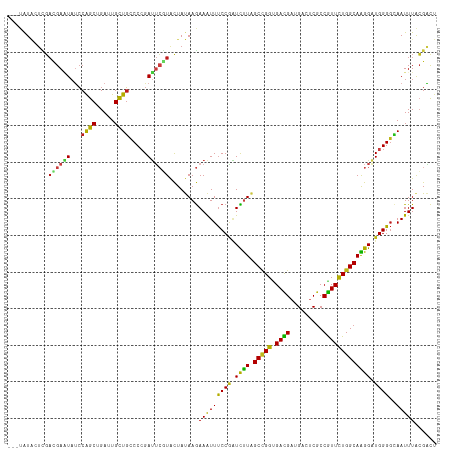
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:49:40 2011