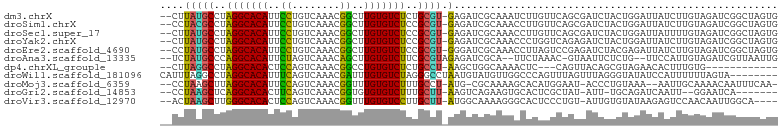
| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,375,989 – 16,376,089 |
| Length | 100 |
| Max. P | 0.999920 |

| Location | 16,375,989 – 16,376,089 |
|---|---|
| Length | 100 |
| Sequences | 11 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 61.55 |
| Shannon entropy | 0.85023 |
| G+C content | 0.46320 |
| Mean single sequence MFE | -25.67 |
| Consensus MFE | -9.70 |
| Energy contribution | -9.40 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.972205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
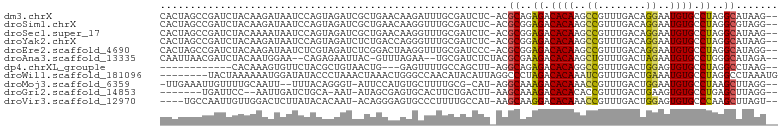
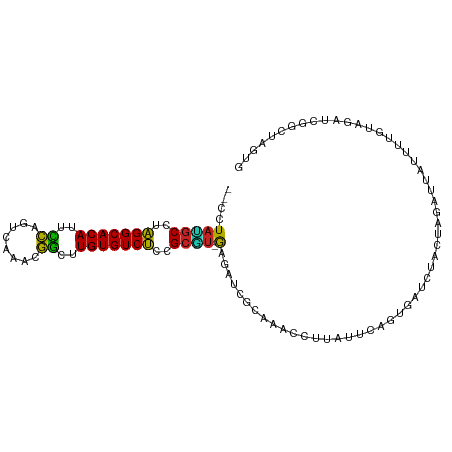
>dm3.chrX 16375989 100 + 22422827 CACUAGCCGAUCUACAAGAUAAUCCAGUAGAUCGCUGAACAAGAUUUGCGAUCUC-ACGCAGAGACACAAGCCGUUUGACAGGAAUGUGCCUAGGCAUAAG-- .....((((((((((..(.....)..)))))))...........((((((.....-.))))))(.((((..((........))..)))).)..))).....-- ( -27.00, z-score = -2.34, R) >droSim1.chrX 12587498 100 + 17042790 CACUAGCCGAUCUACAAGAUAAUCCAGUAGAUCGCUGAACAAGGUUUGCGAUCUC-ACGCGGAGACACAAGCCGUUUGACAGGAAUGUGCCUAGGCGUAGG-- ..(((((((((((((..(.....)..)))))))(..((((..((((((.(.((((-.....))))).))))))))))..)(((......))).))).))).-- ( -31.90, z-score = -2.24, R) >droSec1.super_17 195526 100 + 1527944 CACUAGCCGAUCUACAAAAUAAUCCAGUAGAUCGCUGAACAAGGUUUGCGAUCUC-ACGCGGAGACACAAGCCGUUUGACAGGAAUGUGCCUAGGCAUAAG-- ...((((.(((((((...........))))))))))).....((((((.(.((((-.....))))).))))))............(((((....)))))..-- ( -30.50, z-score = -2.90, R) >droYak2.chrX 10494941 100 + 21770863 CACUAGCCGAUCUACAAGAUAAUCCAGUAGAUCUCUGACCAGGGUUUGCGAUCUC-ACGCGGAGACACAAGCCGUUUGACAGGAAUGUGCCUAGGCAUAAG-- .....((((((((((..(.....)..)))))))((((...(.((((((.(.((((-.....))))).)))))).)....))))..........))).....-- ( -28.80, z-score = -1.58, R) >droEre2.scaffold_4690 8026642 100 - 18748788 CACUAGCCGAUCUACAAGAUAAUCUCGUAGAUCUCGGACUAAGGUUUGCGAUCCC-ACGCGGAGACACAAGCCGUUUGACAGGAAUGUGCCUAGGCAUAGG-- ..(((((((((((((.((.....)).)))))))((((((...((((((.(.((((-....)).))).)))))))))))).(((......))).))).))).-- ( -33.40, z-score = -2.73, R) >droAna3.scaffold_13335 1773120 96 - 3335858 CAAUUAACGAUCUACAAUGGAA--CAGAGAAUUAC-GUUUAGAA--UGCGAUCUCUACGCGAAGACACAAGCUGUUUGACUAGAAUGUGCCUGGGCAUAGA-- ..........((((...(.(((--(((.......(-((.((((.--.......)))).)))..........)))))).).)))).(((((....)))))..-- ( -17.53, z-score = 0.08, R) >dp4.chrXL_group1e 9478425 85 - 12523060 ------------CACAAAGUGUUCUACGCUGUAACUG---GAGUUUUGCCAGCUU-AGGCAGAGACACAGGCCGUUUGACUGGAGUGUGCCUAGGCCUAAG-- ------------.....((((.....))))....(((---(.......))))(((-((((..((.((((..(((......)))..)))).))..)))))))-- ( -27.60, z-score = -0.74, R) >droWil1.scaffold_181096 5794900 95 + 12416693 --------UACUAAAAAAUGGAUAUACCCUAAACUAAACUGGGCCAACAUACAUUAGGCCCUAGACACAAAUCGUUUGACUGAAAUGUGCCUAGGCCUAAAUG --------..........(((.....(((...........)))))).......(((((((.(((.((((..(((......)))..)))).))))))))))... ( -22.50, z-score = -3.19, R) >droMoj3.scaffold_6359 3297206 95 + 4525533 -UUGAAAUUGUUUUGCAAUU--UUUACAGGGU-AUUCCAUGUGCUUUUGCG-CAU-AGGCAAAGACACAAACCGUUUGACUGGAAUGUGCCUAAGCUUAGG-- -..(((((((.....)))))--)).....((.-...)).(((((....)))-))(-((((..((.((((..(((......)))..)))).))..)))))..-- ( -22.30, z-score = -0.33, R) >droGri2.scaffold_14853 3232060 89 - 10151454 -------UGAUUCC--AAUUGAUCUGCA-AAU-AUAGCGAGUGCACUUCUGACUU-AAGCAAAGACACACACCGUUUGACUGAAGUGUGCCUGAGCUUAGG-- -------.......--....((..((((-...-........))))..))...(((-((((..((.(((((..((......))..))))).))..)))))))-- ( -18.90, z-score = -0.24, R) >droVir3.scaffold_12970 2083277 95 + 11907090 ----UGCCAAUUGUUGGACUCUUAUACACAAU-ACAGGGAGUGCCCUUUUGCCAU-AAGCAAGGACACAAACCGUUUGACUGGAGUGUGCCCAAGCUUAGU-- ----((.(((..(..((((((((.........-...)))))).)))..))).))(-((((..((.((((..(((......)))..)))).))..)))))..-- ( -21.90, z-score = 0.49, R) >consensus CACUAGCCGAUCUACAAAAUAAUCUAGUAGAUCACUGAACAAGCUUUGCGAUCUC_ACGCAGAGACACAAGCCGUUUGACUGGAAUGUGCCUAGGCAUAGG__ ..........................................................((..((.((((..((........))..)))).))..))....... ( -9.70 = -9.40 + -0.30)
| Location | 16,375,989 – 16,376,089 |
|---|---|
| Length | 100 |
| Sequences | 11 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 61.55 |
| Shannon entropy | 0.85023 |
| G+C content | 0.46320 |
| Mean single sequence MFE | -30.25 |
| Consensus MFE | -15.26 |
| Energy contribution | -14.47 |
| Covariance contribution | -0.79 |
| Combinations/Pair | 1.54 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.90 |
| SVM RNA-class probability | 0.999920 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
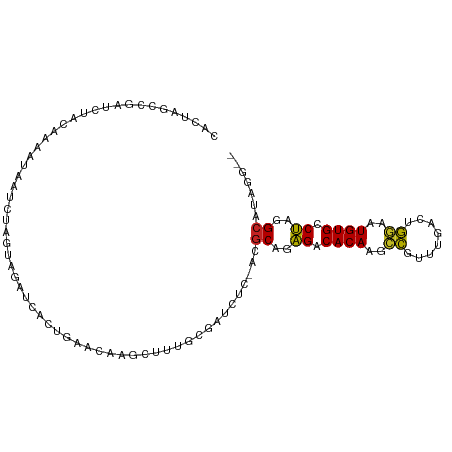
>dm3.chrX 16375989 100 - 22422827 --CUUAUGCCUAGGCACAUUCCUGUCAAACGGCUUGUGUCUCUGCGU-GAGAUCGCAAAUCUUGUUCAGCGAUCUACUGGAUUAUCUUGUAGAUCGGCUAGUG --(((((((..(((((((..((........))..)))))))..))))-)))................((((((((((.((.....)).))))))).))).... ( -34.30, z-score = -3.29, R) >droSim1.chrX 12587498 100 - 17042790 --CCUACGCCUAGGCACAUUCCUGUCAAACGGCUUGUGUCUCCGCGU-GAGAUCGCAAACCUUGUUCAGCGAUCUACUGGAUUAUCUUGUAGAUCGGCUAGUG --..(((((..(((((((..((........))..)))))))..))))-)(((((((............)))))))(((((...(((.....)))...))))). ( -33.30, z-score = -2.73, R) >droSec1.super_17 195526 100 - 1527944 --CUUAUGCCUAGGCACAUUCCUGUCAAACGGCUUGUGUCUCCGCGU-GAGAUCGCAAACCUUGUUCAGCGAUCUACUGGAUUAUUUUGUAGAUCGGCUAGUG --(((((((..(((((((..((........))..)))))))..))))-)))................((((((((((...........))))))).))).... ( -32.10, z-score = -2.60, R) >droYak2.chrX 10494941 100 - 21770863 --CUUAUGCCUAGGCACAUUCCUGUCAAACGGCUUGUGUCUCCGCGU-GAGAUCGCAAACCCUGGUCAGAGAUCUACUGGAUUAUCUUGUAGAUCGGCUAGUG --(((((((..(((((((..((........))..)))))))..))))-)))..........((((((...(((((((.((.....)).))))))))))))).. ( -36.50, z-score = -3.32, R) >droEre2.scaffold_4690 8026642 100 + 18748788 --CCUAUGCCUAGGCACAUUCCUGUCAAACGGCUUGUGUCUCCGCGU-GGGAUCGCAAACCUUAGUCCGAGAUCUACGAGAUUAUCUUGUAGAUCGGCUAGUG --(((((((..(((((((..((........))..)))))))..))))-)))..........((((((...((((((((((.....)))))))))))))))).. ( -40.40, z-score = -4.55, R) >droAna3.scaffold_13335 1773120 96 + 3335858 --UCUAUGCCCAGGCACAUUCUAGUCAAACAGCUUGUGUCUUCGCGUAGAGAUCGCA--UUCUAAAC-GUAAUUCUCUG--UUCCAUUGUAGAUCGUUAAUUG --(((((((..(((((((..((........))..)))))))..)))))))(((((((--.....(((-(........))--))....))).))))........ ( -24.20, z-score = -3.00, R) >dp4.chrXL_group1e 9478425 85 + 12523060 --CUUAGGCCUAGGCACACUCCAGUCAAACGGCCUGUGUCUCUGCCU-AAGCUGGCAAAACUC---CAGUUACAGCGUAGAACACUUUGUG------------ --(((((((..(((((((..((........))..)))))))..))))-)))((((.......)---))).(((((.((.....)).)))))------------ ( -26.10, z-score = -1.53, R) >droWil1.scaffold_181096 5794900 95 - 12416693 CAUUUAGGCCUAGGCACAUUUCAGUCAAACGAUUUGUGUCUAGGGCCUAAUGUAUGUUGGCCCAGUUUAGUUUAGGGUAUAUCCAUUUUUUAGUA-------- ...(((((((((((((((..((........))..)))))))).)))))))((.((((..((((...........)))))))).))..........-------- ( -29.50, z-score = -3.51, R) >droMoj3.scaffold_6359 3297206 95 - 4525533 --CCUAAGCUUAGGCACAUUCCAGUCAAACGGUUUGUGUCUUUGCCU-AUG-CGCAAAAGCACAUGGAAU-ACCCUGUAAA--AAUUGCAAAACAAUUUCAA- --.....((..(((((((..((........))..)))))))..))((-(((-.((....)).)))))...-.........(--(((((.....))))))...- ( -24.50, z-score = -2.34, R) >droGri2.scaffold_14853 3232060 89 + 10151454 --CCUAAGCUCAGGCACACUUCAGUCAAACGGUGUGUGUCUUUGCUU-AAGUCAGAAGUGCACUCGCUAU-AUU-UGCAGAUCAAUU--GGAAUCA------- --(((((((..((((((((.((........)).))))))))..))))-).....((..((((........-...-))))..))....--)).....------- ( -22.30, z-score = -0.78, R) >droVir3.scaffold_12970 2083277 95 - 11907090 --ACUAAGCUUGGGCACACUCCAGUCAAACGGUUUGUGUCCUUGCUU-AUGGCAAAAGGGCACUCCCUGU-AUUGUGUAUAAGAGUCCAACAAUUGGCA---- --..(((((..(((((((..((........))..)))))))..))))-)((.(((...((.((((..(((-((...))))).)))))).....))).))---- ( -29.50, z-score = -1.44, R) >consensus __CCUAUGCCUAGGCACAUUCCAGUCAAACGGCUUGUGUCUCCGCGU_GAGAUCGCAAACCUUAUUCAGUGAUCUACUAGAUUAUUUUGUAGAUCGGCUAGUG .....((((..(((((((..((........))..)))))))..))))........................................................ (-15.26 = -14.47 + -0.79)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:49:38 2011