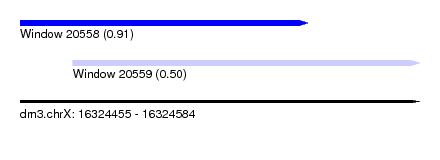
| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,324,455 – 16,324,584 |
| Length | 129 |
| Max. P | 0.906811 |

| Location | 16,324,455 – 16,324,548 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 95.81 |
| Shannon entropy | 0.07444 |
| G+C content | 0.44063 |
| Mean single sequence MFE | -24.36 |
| Consensus MFE | -22.80 |
| Energy contribution | -23.20 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.906811 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
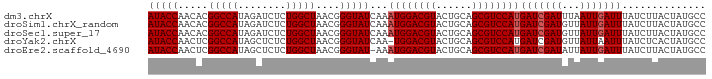
>dm3.chrX 16324455 93 + 22422827 AUACCAACACGGCCAUAGAUCUCUGGCUAACGGGUAUCAAAUGGACGUACUGCAGCGUCCAUGAUCGAUUUAAUUGAUUUAUCUUACUAUGCC (((((.....(((((........)))))....)))))...((((((((......))))))))(((((((...))))))).............. ( -24.40, z-score = -2.16, R) >droSim1.chrX_random 4291015 93 + 5698898 AUACCAACACGGCCAUAGAUCUCUGGCUAACGGGUAUCAAAUGGACGUACUGCAGCGUCCAUGAUCGAUGUUAUUGAUUUAUCUUACUAUGCC (((((.....(((((........)))))....)))))...((((((((......))))))))(((((((...))))))).............. ( -24.40, z-score = -1.58, R) >droSec1.super_17 144143 93 + 1527944 AUACCAACACGGCCAUAGAUCUCUGGCUAACGGGUAUCAAAUGGACGUACUGCAGCGUCCAUGAUCGAUGUUAUUGAUUUAUCUUACUAUGCC (((((.....(((((........)))))....)))))...((((((((......))))))))(((((((...))))))).............. ( -24.40, z-score = -1.58, R) >droYak2.chrX 10443833 92 + 21770863 AUACCAACUCGGCCAUAGCUCUCUGGCUAACGGGUAUCAA-UGGACGUACUGCAGCGUCCAUGAUCGAUGUUAUUAAUUUAUCUCACUAUGCC .....(((((((((.(((((....)))))...)))((((.-(((((((......)))))))))))))).)))..................... ( -23.60, z-score = -1.58, R) >droEre2.scaffold_4690 7975493 92 - 18748788 AUACCAACUCGGCCAUAGCUCUCUGGCUAACGGGUAU-AAAUGGACGUACUGCAGCGUCCAUGAUCGAUAUUAUUGAUUUAUCUUACUAUGCC (((((.....(((((........)))))....)))))-..((((((((......))))))))(((((((...))))))).............. ( -25.00, z-score = -2.20, R) >consensus AUACCAACACGGCCAUAGAUCUCUGGCUAACGGGUAUCAAAUGGACGUACUGCAGCGUCCAUGAUCGAUGUUAUUGAUUUAUCUUACUAUGCC (((((.....(((((........)))))....)))))...((((((((......))))))))(((((((...))))))).............. (-22.80 = -23.20 + 0.40)
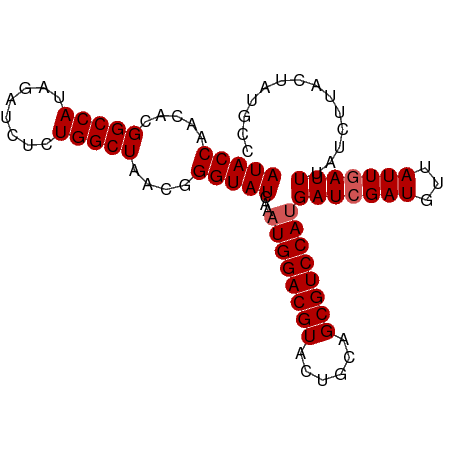
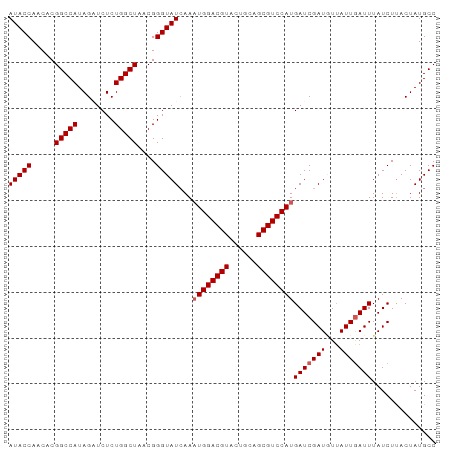
| Location | 16,324,472 – 16,324,584 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 94.18 |
| Shannon entropy | 0.10071 |
| G+C content | 0.37367 |
| Mean single sequence MFE | -26.38 |
| Consensus MFE | -22.68 |
| Energy contribution | -23.48 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chrX 16324472 112 + 22422827 GAUCUCUGGCUAACGGGUAUCAAAUGGACGUACUGCAGCGUCCAUGAUCGAUUUAAUUGAUUUAUCUUACUAUGCCAUUUACGAAUUUGUGCAAAUGUUGUAUAUUCGAUUU ......((((.....((((....((((((((......))))))))(((((((...))))))).....))))..))))....(((((..(((((.....)))))))))).... ( -27.80, z-score = -1.93, R) >droSim1.chrX_random 4291032 112 + 5698898 GAUCUCUGGCUAACGGGUAUCAAAUGGACGUACUGCAGCGUCCAUGAUCGAUGUUAUUGAUUUAUCUUACUAUGCCAUUUACGAAUUUGUGCAAAUGUUGUUUAUUCGAUUU ......((((.....((((....((((((((......))))))))(((((((...))))))).....))))..))))....(((((....(((.....)))..))))).... ( -26.20, z-score = -1.13, R) >droSec1.super_17 144160 112 + 1527944 GAUCUCUGGCUAACGGGUAUCAAAUGGACGUACUGCAGCGUCCAUGAUCGAUGUUAUUGAUUUAUCUUACUAUGCCAUUUACGAAUUUGUGCAAAUGUUGUUUAUUCGAUUU ......((((.....((((....((((((((......))))))))(((((((...))))))).....))))..))))....(((((....(((.....)))..))))).... ( -26.20, z-score = -1.13, R) >droYak2.chrX 10443850 109 + 21770863 GCUCUCUGGCUAACGGGUAU-CAAUGGACGUACUGCAGCGUCCAUGAUCGAUGUUAUUAAUUUAUCUCACUAUGCCAUUUACGAACUUG--CAAAUGUUGUAUAUUCGAUAU ......((((.....(((((-((.(((((((......))))))))))).((((.........))))..)))..))))....((((..((--((.....))))..)))).... ( -26.20, z-score = -1.51, R) >droEre2.scaffold_4690 7975510 109 - 18748788 GCUCUCUGGCUAACGGGUAU-AAAUGGACGUACUGCAGCGUCCAUGAUCGAUAUUAUUGAUUUAUCUUACUAUGCCAUUUACGAAUUUU--CAAAUGUUGUAUAUUCGAUUU (((....))).....(((((-(.((((((((......))))))))(((((((...)))))))........)))))).....(((((...--((.....))...))))).... ( -25.50, z-score = -2.01, R) >consensus GAUCUCUGGCUAACGGGUAUCAAAUGGACGUACUGCAGCGUCCAUGAUCGAUGUUAUUGAUUUAUCUUACUAUGCCAUUUACGAAUUUGUGCAAAUGUUGUAUAUUCGAUUU ......((((....(((((....((((((((......))))))))(((((((...))))))))))))......))))....(((((....(((.....)))..))))).... (-22.68 = -23.48 + 0.80)

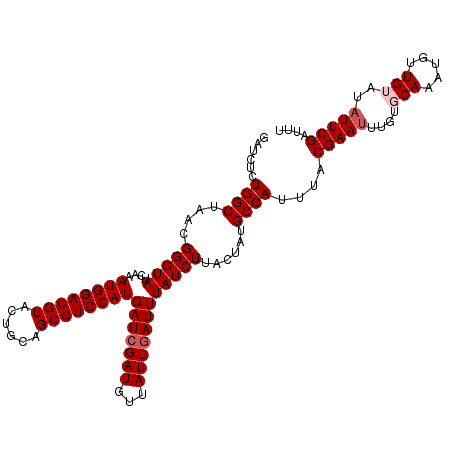
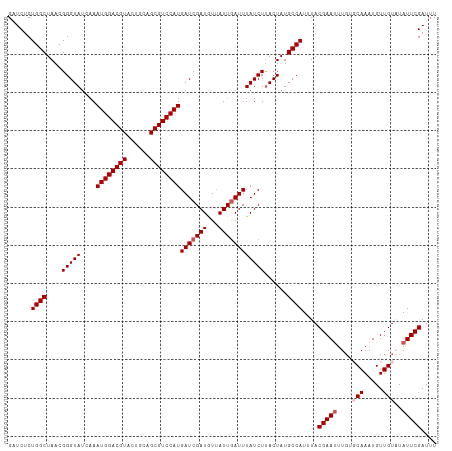
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:49:34 2011