| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,322,563 – 16,322,624 |
| Length | 61 |
| Max. P | 0.865767 |

| Location | 16,322,563 – 16,322,624 |
|---|---|
| Length | 61 |
| Sequences | 8 |
| Columns | 71 |
| Reading direction | reverse |
| Mean pairwise identity | 70.27 |
| Shannon entropy | 0.56172 |
| G+C content | 0.46189 |
| Mean single sequence MFE | -18.48 |
| Consensus MFE | -8.66 |
| Energy contribution | -7.86 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.83 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.865767 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
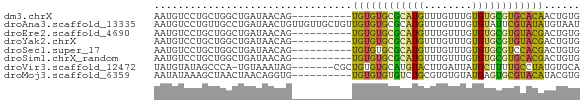
>dm3.chrX 16322563 61 - 22422827 AAUGUCCUGCUGGCUGAUAACAG----------UGUGUGCGCAUGUUUGUUUGUGUGCGUGCACAACUGUG ..((((..(....).))))((((----------((((..((((..(......)..))))..))).))))). ( -22.90, z-score = -2.42, R) >droAna3.scaffold_13335 1477333 71 - 3335858 AAUGUCCUGUUGCCUGAUAACUGUUGUUGCUGUUGUGUGCGCAUGUUUGUUUGUGUAUUCGUAUAUGUAAU ........(((((...(((((.((....)).)))))(((((((........)))))))........))))) ( -11.30, z-score = -0.61, R) >droEre2.scaffold_4690 7973547 61 + 18748788 AAUGUCCUGCUGGCUGAUAACAG----------UGUGUGCGCAUGUUUGUUUGUGUGCGUGUACGACUGUG ..((((..(....).))))((((----------((((..((((..(......)..))))..))).))))). ( -20.40, z-score = -2.25, R) >droYak2.chrX 10441895 61 - 21770863 AAUGUCCUGCUGGCUGAUAACAG----------UGUGUGCGCAUGUUUGUUUGUGUGCGUGUACGACUGUG ..((((..(....).))))((((----------((((..((((..(......)..))))..))).))))). ( -20.40, z-score = -2.25, R) >droSec1.super_17 142294 61 - 1527944 AAUGUCCUGCUGGCUGAUAACAG----------UGUGUGCGCAUGUUUGUUUGUGUGCGUCCACGACUGUG ..((((..(....).))))((((----------((((..((((..(......)..))))..))).))))). ( -19.90, z-score = -1.95, R) >droSim1.chrX_random 4289348 61 - 5698898 AAUGUCCUGCUGGCUGAUAACAG----------UGUGUGCGCAUGUUUGUUUGUGUGCGUGCACGACUGUG ..((((..(....).))))((((----------((((..((((..(......)..))))..))).))))). ( -22.90, z-score = -2.52, R) >droVir3.scaffold_12472 574665 63 + 763072 UAUGUAUAGCCCA-UGUAAAUAG-------CGCUGUGUGCAUGUACUUGAUUAUGCUUUUGCCUAUGUGCA .............-((((.((((-------.((.....(((((........)))))....)))))).)))) ( -12.70, z-score = 0.47, R) >droMoj3.scaffold_6359 2690434 61 - 4525533 AAUAUAAAGCUAACUAACAGGUG----------UGUGUGUGUCUGCGUGUGUAUGAGUGCGUACAUACGUG .(((((..(((........))).----------..)))))....(((((((((((....))))))))))). ( -17.30, z-score = -1.02, R) >consensus AAUGUCCUGCUGGCUGAUAACAG__________UGUGUGCGCAUGUUUGUUUGUGUGCGUGCACAACUGUG .................................((((((((((((........))))))))))))...... ( -8.66 = -7.86 + -0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:49:32 2011