
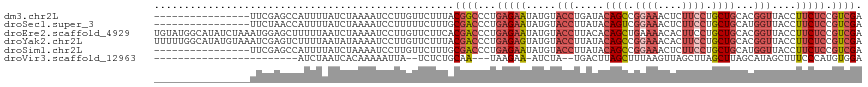
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,511,002 – 11,511,144 |
| Length | 142 |
| Max. P | 0.991387 |

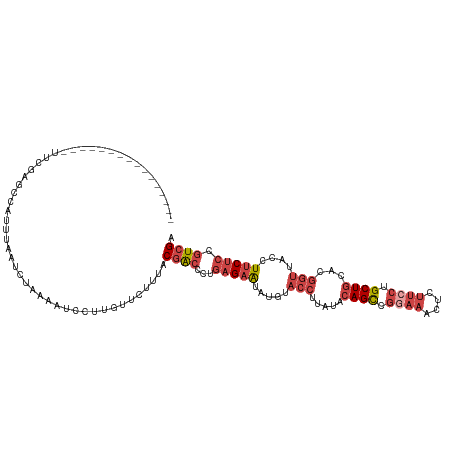
| Location | 11,511,002 – 11,511,104 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 68.58 |
| Shannon entropy | 0.56419 |
| G+C content | 0.40859 |
| Mean single sequence MFE | -21.62 |
| Consensus MFE | -12.43 |
| Energy contribution | -14.35 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.24 |
| SVM RNA-class probability | 0.986452 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 11511002 102 + 23011544 ----------------UUCGAGCCAUUUUAUCUAAAAUCCUUGUUCUUUACGGCCCUGAGAAUAUGUACCUGAUACAGCCGGAAACUCUUCCUGCUGCACGGUUACCUUCUCCGUCGA ----------------...((((.(((((....)))))....))))....((((...(((((...(((.(((...((((.((((....)))).))))..))).))).))))).)))). ( -23.70, z-score = -2.08, R) >droSec1.super_3 6908149 102 + 7220098 ----------------UUCUAACCAUUUUAUCUAAAAUCCUUUUUCUUUGCGACCCUGAGAAUAUGUACCUUAUACAGUCGGAAACUCUUCCUGCUGCAUGGUUACCUUCUCCGUCGA ----------------..................................((((...(((((.....(((.....((((.((((....)))).))))...)))....))))).)))). ( -19.80, z-score = -2.74, R) >droEre2.scaffold_4929 12719991 118 - 26641161 UGUAUGGCAUAUCUAAAUGGAGCUUUUUAAUCUAAAAUCCUUGUUCUUCACGACCCUGAGAAUAUGUACCUUACACAGCUGAAAACACUUCCUGCUGCACGGUUACCUUCUCCGUCGA ..................(((((...................)))))...((((...(((((.....(((.....((((.(((.....)))..))))...)))....))))).)))). ( -19.01, z-score = 0.12, R) >droYak2.chr2L 7917597 118 + 22324452 UUUUUGGCAUAUGUAAAUCGAGUCUUUUAAUAUAAAAUCCUUGUUCUUUACGACCCUGAGAGUAUGUACCUUAUACAGCCGGAAACACUUCCUGCUGCACGGUUACCUUCUCCGUCGA ...(((((...((((((.((((..(((((...)))))..))))...)))))).....(((((.....(((.....((((.((((....)))).))))...)))....))))).))))) ( -26.00, z-score = -1.78, R) >droSim1.chr2L 11326923 102 + 22036055 ----------------UUCGAGCCAUUUUAUCUAAAAUCCUUGUUCUUUGCGACCCUGAGAAUAUGUACCUUAUACAGCCGGAAACUCUUCCUGCUGCAUGGUUACCUUCUCCGUCGA ----------------...((((.(((((....)))))....))))....((((...(((((.....(((.....((((.((((....)))).))))...)))....))))).)))). ( -23.10, z-score = -2.70, R) >droVir3.scaffold_12963 18169997 86 - 20206255 ------------------------AUCUAAUCACAAAAAUUA--UCUCUGCAA---UAAGAA-AUCUA--UGACUUAGCUUUAAGUUAGCUUAGCUUAGCAUAGCUUUCCCAUGUGGA ------------------------..................--..((..((.---...(((-(.(((--((.((.((((..(((....))))))).))))))).))))...))..)) ( -18.10, z-score = -1.49, R) >consensus ________________UUCGAGCCAUUUAAUCUAAAAUCCUUGUUCUUUACGACCCUGAGAAUAUGUACCUUAUACAGCCGGAAACUCUUCCUGCUGCACGGUUACCUUCUCCGUCGA ..................................................((((...(((((.....(((.....((((.((((....)))).))))...)))....))))).)))). (-12.43 = -14.35 + 1.92)
| Location | 11,511,002 – 11,511,104 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 68.58 |
| Shannon entropy | 0.56419 |
| G+C content | 0.40859 |
| Mean single sequence MFE | -26.20 |
| Consensus MFE | -17.65 |
| Energy contribution | -18.60 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.47 |
| SVM RNA-class probability | 0.991387 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 11511002 102 - 23011544 UCGACGGAGAAGGUAACCGUGCAGCAGGAAGAGUUUCCGGCUGUAUCAGGUACAUAUUCUCAGGGCCGUAAAGAACAAGGAUUUUAGAUAAAAUGGCUCGAA---------------- ((....(((((.(((.(((((((((.((((....)))).)))))))..)))))...))))).(((((((.......................))))))))).---------------- ( -31.30, z-score = -2.73, R) >droSec1.super_3 6908149 102 - 7220098 UCGACGGAGAAGGUAACCAUGCAGCAGGAAGAGUUUCCGACUGUAUAAGGUACAUAUUCUCAGGGUCGCAAAGAAAAAGGAUUUUAGAUAAAAUGGUUAGAA---------------- .((((.(((((.(((.((((((((..((((....))))..))))))..)))))...)))))...))))..................................---------------- ( -24.40, z-score = -2.28, R) >droEre2.scaffold_4929 12719991 118 + 26641161 UCGACGGAGAAGGUAACCGUGCAGCAGGAAGUGUUUUCAGCUGUGUAAGGUACAUAUUCUCAGGGUCGUGAAGAACAAGGAUUUUAGAUUAAAAAGCUCCAUUUAGAUAUGCCAUACA .((((.(((((.(((.(((..((((.((((....)))).))))..)..)))))...)))))...))))..........((..(((((((...........)))))))....))..... ( -25.50, z-score = -0.11, R) >droYak2.chr2L 7917597 118 - 22324452 UCGACGGAGAAGGUAACCGUGCAGCAGGAAGUGUUUCCGGCUGUAUAAGGUACAUACUCUCAGGGUCGUAAAGAACAAGGAUUUUAUAUUAAAAGACUCGAUUUACAUAUGCCAAAAA ..(((.((((..(((.(((((((((.((((....)))).)))))))..)))))....))))...)))(((((...(.((..(((((...)))))..)).).)))))............ ( -29.00, z-score = -1.53, R) >droSim1.chr2L 11326923 102 - 22036055 UCGACGGAGAAGGUAACCAUGCAGCAGGAAGAGUUUCCGGCUGUAUAAGGUACAUAUUCUCAGGGUCGCAAAGAACAAGGAUUUUAGAUAAAAUGGCUCGAA---------------- .((((.(((((.(((.(((((((((.((((....)))).)))))))..)))))...)))))...))))..................................---------------- ( -27.00, z-score = -2.10, R) >droVir3.scaffold_12963 18169997 86 + 20206255 UCCACAUGGGAAAGCUAUGCUAAGCUAAGCUAACUUAAAGCUAAGUCA--UAGAU-UUCUUA---UUGCAGAGA--UAAUUUUUGUGAUUAGAU------------------------ ......(((((((.(((((((.(((((((....)))..)))).)).))--))).)-))))))---(..((((((--...))))))..)......------------------------ ( -20.00, z-score = -1.24, R) >consensus UCGACGGAGAAGGUAACCGUGCAGCAGGAAGAGUUUCCGGCUGUAUAAGGUACAUAUUCUCAGGGUCGCAAAGAACAAGGAUUUUAGAUAAAAUGGCUCGAA________________ .((((.(((((.(((.(((((((((.(((......))).)))))))..)))))...)))))...)))).................................................. (-17.65 = -18.60 + 0.95)

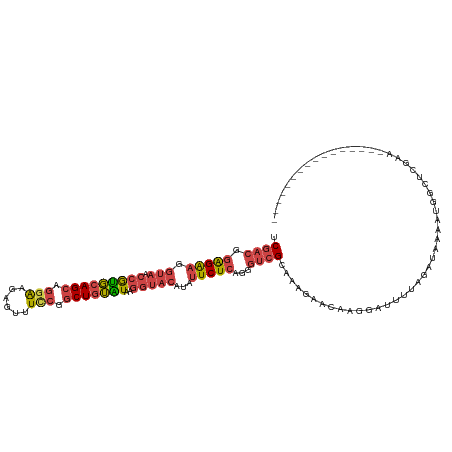
| Location | 11,511,026 – 11,511,133 |
|---|---|
| Length | 107 |
| Sequences | 7 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 64.88 |
| Shannon entropy | 0.70716 |
| G+C content | 0.46052 |
| Mean single sequence MFE | -27.40 |
| Consensus MFE | -10.41 |
| Energy contribution | -10.47 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.916954 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 11511026 107 + 23011544 UUGUUCUUUACGGCCCUGAGAAUAUGUACCUGAUACAGCCGGAAACUCUUCCUGCUGCACGGUUACCUUCUCCGUCGAA---GGCACGAAUUAUGGGGAAAUGCUACUAU ((((.((((.((((...(((((...(((.(((...((((.((((....)))).))))..))).))).))))).))))))---)).))))..................... ( -26.70, z-score = -0.67, R) >droAna3.scaffold_12916 9231460 105 - 16180835 UCGUUAGUGGGGACC-----AGUCUCUACCUGACACAGCUGGAAGCGAUUCCCGCUGCACUUCUUCACGGUGGACGGGGAUUGACACAGGGAAUGGGGUUUAGUAACUAG ..((((.((((..((-----(.(((((.((..........))..((((((((((.(.((((.......)))).)))))))))).)..))))).)))..)))).))))... ( -34.10, z-score = -0.63, R) >droSec1.super_3 6908173 107 + 7220098 UUUUUCUUUGCGACCCUGAGAAUAUGUACCUUAUACAGUCGGAAACUCUUCCUGCUGCAUGGUUACCUUCUCCGUCGAA---GGCAUGAACUAUGGGGACAUGCUACUAC ...(((((.........)))))(((((.(((....((((.((((....)))).))))(((((((.(((((......)))---))....)))))))))))))))....... ( -28.20, z-score = -1.97, R) >droEre2.scaffold_4929 12720031 107 - 26641161 UUGUUCUUCACGACCCUGAGAAUAUGUACCUUACACAGCUGAAAACACUUCCUGCUGCACGGUUACCUUCUCCGUCGAA---GGAUCGAACUAUGGGAACAUGCUACUAC ..((((.((.((((...(((((.....(((.....((((.(((.....)))..))))...)))....))))).))))..---.))..))))((((....))))....... ( -22.80, z-score = -1.13, R) >droYak2.chr2L 7917637 107 + 22324452 UUGUUCUUUACGACCCUGAGAGUAUGUACCUUAUACAGCCGGAAACACUUCCUGCUGCACGGUUACCUUCUCCGUCGAA---GGAUCGAACUAUGGGAACAUGCUACUAC ((((.....)))).....(((((((((.((.....((((.((((....)))).))))((..(((.(((((......)))---))....)))..)))).))))))).)).. ( -30.20, z-score = -2.56, R) >droSim1.chr2L 11326947 107 + 22036055 UUGUUCUUUGCGACCCUGAGAAUAUGUACCUUAUACAGCCGGAAACUCUUCCUGCUGCAUGGUUACCUUCUCCGUCGAA---GGCACGAACUAUGGGGACAUGCUACUAC ((((.((((.((((...(((((.....(((.....((((.((((....)))).))))...)))....))))).))))))---)).))))..((((....))))....... ( -30.50, z-score = -2.39, R) >droVir3.scaffold_12963 18170015 98 - 20206255 ----UCUCUGCAAU------AAGAAAUCUAUGACUUAGCUUUAAGUUAGCUUAGCUUAGCAUAGCUUUCCCAUGUGGAACCCACUUAAUUUUAUUAACAAAAUAGAUU-- ----..((..((..------..((((.(((((.((.((((..(((....))))))).))))))).))))...))..))...........((((((.....))))))..-- ( -19.30, z-score = -1.56, R) >consensus UUGUUCUUUGCGACCCUGAGAAUAUGUACCUUAUACAGCCGGAAACUCUUCCUGCUGCACGGUUACCUUCUCCGUCGAA___GGCACGAACUAUGGGGACAUGCUACUAC .............(((...................((((.((((....)))).)))).((((.........))))...................)))............. (-10.41 = -10.47 + 0.06)

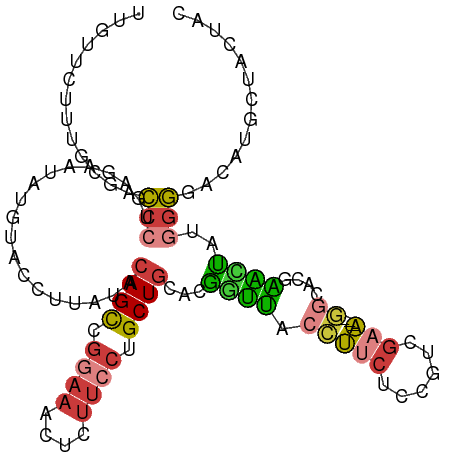
| Location | 11,511,026 – 11,511,133 |
|---|---|
| Length | 107 |
| Sequences | 7 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 64.88 |
| Shannon entropy | 0.70716 |
| G+C content | 0.46052 |
| Mean single sequence MFE | -28.81 |
| Consensus MFE | -13.56 |
| Energy contribution | -14.57 |
| Covariance contribution | 1.01 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.982780 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 11511026 107 - 23011544 AUAGUAGCAUUUCCCCAUAAUUCGUGCC---UUCGACGGAGAAGGUAACCGUGCAGCAGGAAGAGUUUCCGGCUGUAUCAGGUACAUAUUCUCAGGGCCGUAAAGAACAA .............(((........((((---(((......)))))))((((((((((.((((....)))).)))))))..)))...........)))............. ( -29.70, z-score = -1.40, R) >droAna3.scaffold_12916 9231460 105 + 16180835 CUAGUUACUAAACCCCAUUCCCUGUGUCAAUCCCCGUCCACCGUGAAGAAGUGCAGCGGGAAUCGCUUCCAGCUGUGUCAGGUAGAGACU-----GGUCCCCACUAACGA ((((((.((..(((...(((.(.(((............))).).)))((...(((((.((((....)))).))))).)).))))).))))-----))............. ( -22.30, z-score = 0.70, R) >droSec1.super_3 6908173 107 - 7220098 GUAGUAGCAUGUCCCCAUAGUUCAUGCC---UUCGACGGAGAAGGUAACCAUGCAGCAGGAAGAGUUUCCGACUGUAUAAGGUACAUAUUCUCAGGGUCGCAAAGAAAAA ......(((((..(.....)..)))))(---((((((.(((((.(((.((((((((..((((....))))..))))))..)))))...)))))...))))..)))..... ( -30.80, z-score = -2.43, R) >droEre2.scaffold_4929 12720031 107 + 26641161 GUAGUAGCAUGUUCCCAUAGUUCGAUCC---UUCGACGGAGAAGGUAACCGUGCAGCAGGAAGUGUUUUCAGCUGUGUAAGGUACAUAUUCUCAGGGUCGUGAAGAACAA ...((..((((..(((...((.(((...---.))))).(((((.(((.(((..((((.((((....)))).))))..)..)))))...))))).))).))))....)).. ( -30.20, z-score = -1.63, R) >droYak2.chr2L 7917637 107 - 22324452 GUAGUAGCAUGUUCCCAUAGUUCGAUCC---UUCGACGGAGAAGGUAACCGUGCAGCAGGAAGUGUUUCCGGCUGUAUAAGGUACAUACUCUCAGGGUCGUAAAGAACAA .........(((((........((((((---(.....((((...(((.(((((((((.((((....)))).)))))))..)))))...)))).)))))))....))))). ( -32.40, z-score = -2.07, R) >droSim1.chr2L 11326947 107 - 22036055 GUAGUAGCAUGUCCCCAUAGUUCGUGCC---UUCGACGGAGAAGGUAACCAUGCAGCAGGAAGAGUUUCCGGCUGUAUAAGGUACAUAUUCUCAGGGUCGCAAAGAACAA ......(((((..(.....)..)))))(---((((((.(((((.(((.(((((((((.((((....)))).)))))))..)))))...)))))...))))..)))..... ( -33.00, z-score = -2.32, R) >droVir3.scaffold_12963 18170015 98 + 20206255 --AAUCUAUUUUGUUAAUAAAAUUAAGUGGGUUCCACAUGGGAAAGCUAUGCUAAGCUAAGCUAACUUAAAGCUAAGUCAUAGAUUUCUU------AUUGCAGAGA---- --......((((((............((((...))))((((((((.(((((((.(((((((....)))..)))).)).))))).))))))------)).)))))).---- ( -23.30, z-score = -1.46, R) >consensus GUAGUAGCAUGUCCCCAUAGUUCGUGCC___UUCGACGGAGAAGGUAACCGUGCAGCAGGAAGAGUUUCCGGCUGUAUAAGGUACAUAUUCUCAGGGUCGCAAAGAACAA .................................((((.(((((.(((...(((((((.((((....)))).)))))))....)))...)))))...)))).......... (-13.56 = -14.57 + 1.01)
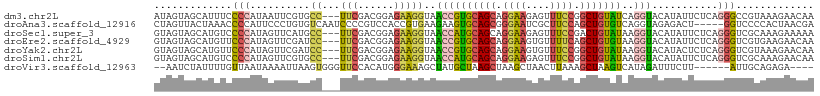

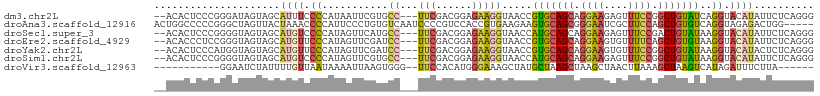
| Location | 11,511,039 – 11,511,144 |
|---|---|
| Length | 105 |
| Sequences | 7 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 63.94 |
| Shannon entropy | 0.71286 |
| G+C content | 0.49314 |
| Mean single sequence MFE | -31.60 |
| Consensus MFE | -13.29 |
| Energy contribution | -13.44 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.58 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.966035 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 11511039 105 - 23011544 --ACACUCCCGGGAUAGUAGCAUUUCCCCAUAAUUCGUGCC---UUCGACGGAGAAGGUAACCGUGCAGCAGGAAGAGUUUCCGGCUGUAUCAGGUACAUAUUCUCAGGG --.....(((((((..........)))).........((((---(((......)))))))((((((((((.((((....)))).)))))))..)))...........))) ( -33.40, z-score = -1.70, R) >droAna3.scaffold_12916 9231473 105 + 16180835 ACUGGCCCCCGGGCUAGUUACUAAACCCCAUUCCCUGUGUCAAUCCCCGUCCACCGUGAAGAAGUGCAGCGGGAAUCGCUUCCAGCUGUGUCAGGUAGAGACUGG----- (((((((....)))))))...........................((.(((((((.(((......(((((.((((....)))).))))).)))))).).))).))----- ( -30.90, z-score = -0.22, R) >droSec1.super_3 6908186 105 - 7220098 --ACACUCCCGGGGUAGUAGCAUGUCCCCAUAGUUCAUGCC---UUCGACGGAGAAGGUAACCAUGCAGCAGGAAGAGUUUCCGACUGUAUAAGGUACAUAUUCUCAGGG --.....(((((((((......)).))))........((((---(((......)))))))(((((((((..((((....))))..))))))..)))...........))) ( -33.20, z-score = -1.68, R) >droEre2.scaffold_4929 12720044 105 + 26641161 --ACACCCUCCGGGUAGUAGCAUGUUCCCAUAGUUCGAUCC---UUCGACGGAGAAGGUAACCGUGCAGCAGGAAGUGUUUUCAGCUGUGUAAGGUACAUAUUCUCAGGG --((((((...)))).))........(((...((.(((...---.))))).(((((.(((.(((..((((.((((....)))).))))..)..)))))...))))).))) ( -28.90, z-score = -0.49, R) >droYak2.chr2L 7917650 105 - 22324452 --ACACUCCCAUGGUAGUAGCAUGUUCCCAUAGUUCGAUCC---UUCGACGGAGAAGGUAACCGUGCAGCAGGAAGUGUUUCCGGCUGUAUAAGGUACAUACUCUCAGGG --.....(((.((..((((...(((.((....(((....((---(((......))))).))).(((((((.((((....)))).)))))))..)).)))))))..))))) ( -31.00, z-score = -0.94, R) >droSim1.chr2L 11326960 105 - 22036055 --ACACUCCCGGGGUAGUAGCAUGUCCCCAUAGUUCGUGCC---UUCGACGGAGAAGGUAACCAUGCAGCAGGAAGAGUUUCCGGCUGUAUAAGGUACAUAUUCUCAGGG --.....(((((((((......)).))))........((((---(((......)))))))((((((((((.((((....)))).)))))))..)))...........))) ( -35.70, z-score = -1.89, R) >droVir3.scaffold_12963 18170024 91 + 20206255 -----------GGAAUCUAUUUUGUUAAUAAAAUUAAGUGGG--UUCCACAUGGGAAAGCUAUGCUAAGCUAAGCUAACUUAAAGCUAAGUCAUAGAUUUCUUA------ -----------(((((((((((.(((.....))).)))))))--))))...(((((((.(((((((.(((((((....)))..)))).)).))))).)))))))------ ( -28.10, z-score = -4.09, R) >consensus __ACACUCCCGGGGUAGUAGCAUGUCCCCAUAGUUCGUGCC___UUCGACGGAGAAGGUAACCGUGCAGCAGGAAGAGUUUCCGGCUGUAUAAGGUACAUAUUCUCAGGG ...................................................(((((.(((...(((((((.((((....)))).)))))))....)))...))))).... (-13.29 = -13.44 + 0.15)
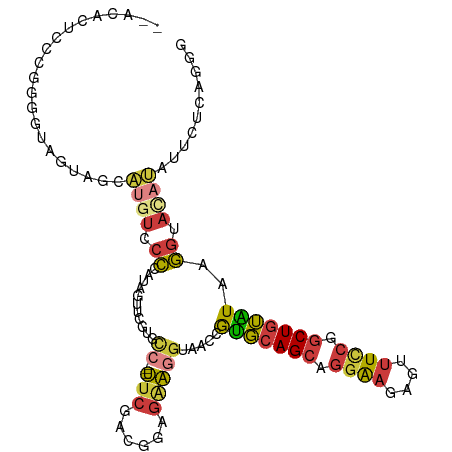
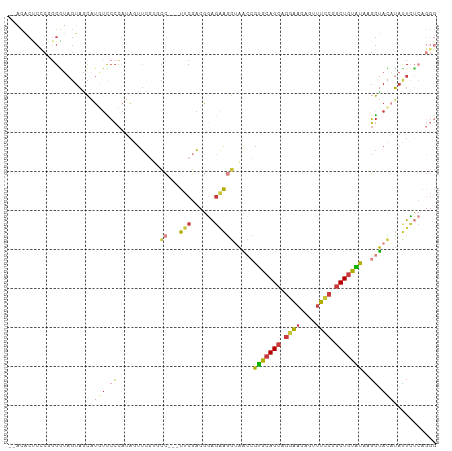
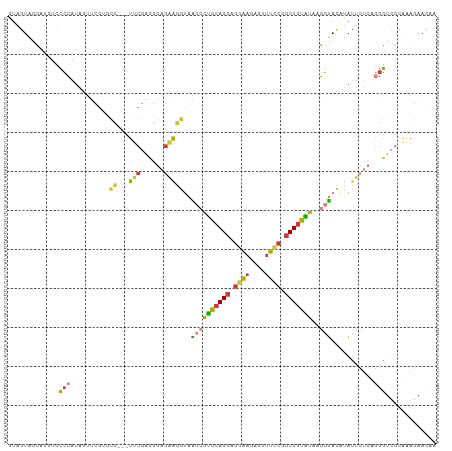
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:33:00 2011