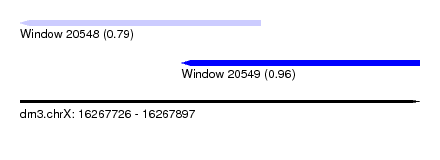
| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,267,726 – 16,267,897 |
| Length | 171 |
| Max. P | 0.960084 |

| Location | 16,267,726 – 16,267,829 |
|---|---|
| Length | 103 |
| Sequences | 7 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 61.42 |
| Shannon entropy | 0.72991 |
| G+C content | 0.37086 |
| Mean single sequence MFE | -15.56 |
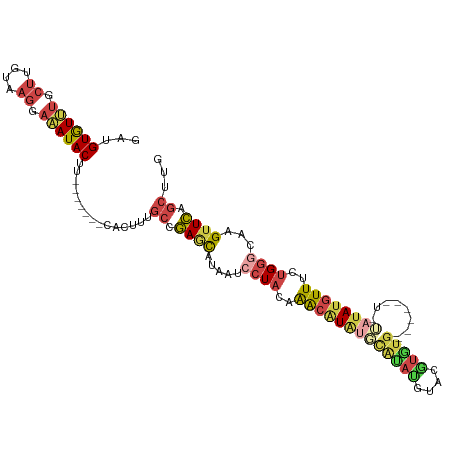
| Consensus MFE | -5.55 |
| Energy contribution | -4.19 |
| Covariance contribution | -1.36 |
| Combinations/Pair | 2.14 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.793698 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
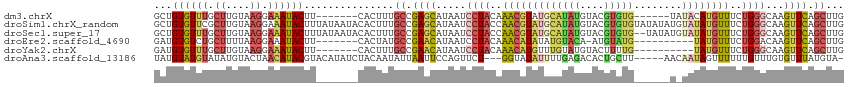
>dm3.chrX 16267726 103 - 22422827 ------GUGUGUAUACAUGUUUCUGGGCAAGUU--CAGCUUGUACAUCUCCCAUUUUCCAUCUCAAUCACUCUCGCUGUAUUUCCCACA-UG-CUUAUUAAAUCACACAUCUC- ------(((((.......((...((((.((((.--((((...................................)))).))))))))..-.)-).........))))).....- ( -17.74, z-score = -2.66, R) >droSim1.chrX_random 4279621 109 - 5698898 GUGUGUAUAUAUGUAUAUGUUUCUGGGCAAGUU--CAGCUUGUACAUCUCCCAUUUUCCAUCUCAGUCACUCUCGCUGUAUUUCCCACA-UG-CUUAUUAAAUCACACAUCUC- (((((.....(((..(((((....(((.((((.--((((..((((....................)).))....)))).))))))))))-))-..))).....))))).....- ( -19.95, z-score = -2.46, R) >droSec1.super_17 84724 107 - 1527944 --GUGUGUAUAUGUAUAUGUUUCUGGGCAAGUU--CAGCUUGUACAUCUCCCAUUUUCCAUCUCAGUCACUCUCGCUGUAUUUCCCACA-UG-CUUAUUAAAUCACACAUCUC- --(((((.....(((...((...((((.((((.--((((..((((....................)).))....)))).))))))))..-.)-).))).....))))).....- ( -19.55, z-score = -2.44, R) >droEre2.scaffold_4690 7918949 99 + 18748788 ----------GUAUGUAUGUUUCUGGACAAGUU--CAGCUUGUACACAUCCCAUUUUCCAUCUCACUUAGUCUCGAAGUAUUUCCCACA-UG-CUUAUUAAAUCACACAUCUC- ----------(((((((((((((.((((((((.--.((..((................)).)).)))).)))).))))))).....)))-))-)...................- ( -11.59, z-score = -0.31, R) >droYak2.chrX 10387422 99 - 21770863 ----------UUUUGUAUGUUUCUGGGCAAGUU--CAGCUUGUACAUCUCCCAUUUUCCACCUCACUCACUCUCGCUGUGAUUCCCACA-CA-CUUAUUUAAUCACACAUCUC- ----------...((((((...(((((....))--)))..))))))............................(.(((((((......-..-.......)))))))).....- ( -11.06, z-score = -0.54, R) >droAna3.scaffold_13186 45413 104 + 49402 -----UGCUUAACAAUAGUUUUUUGUUUGUGUU--UA-UGUAUGCAUAAACGUUUUUGUUUUUCAA-AAUUCGCACUUCGUCGCUCACA-AAUCUUGUUAUCAGAAAAACAUUG -----.((..(((((.......))))).))(((--((-((....))))))).....((((((((..-.....((........))..(((-.....))).....))))))))... ( -16.60, z-score = -0.49, R) >droGri2.scaffold_15203 7002556 105 - 11997470 --------UUACAUACAGACAUACAUAUGUGCUAAUAGCUCACAUGUAUUUGUUUUCUUUUAUUAUAAAUUCACACACGUUCGUUUUUGCCAGCAUUUAAUCACUGAUUUUUU- --------.......(((..((((((.((.((.....)).)).)))))).............................(((.((....)).))).........))).......- ( -12.40, z-score = -0.42, R) >consensus _________UAUAUAUAUGUUUCUGGGCAAGUU__CAGCUUGUACAUCUCCCAUUUUCCAUCUCAAUCACUCUCGCUGUAUUUCCCACA_UG_CUUAUUAAAUCACACAUCUC_ .......................((..(((((.....)))))..)).................................................................... ( -5.55 = -4.19 + -1.36)
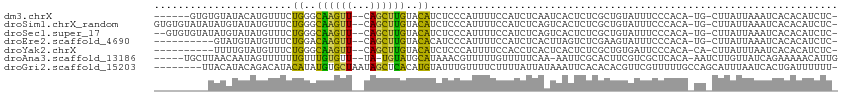
| Location | 16,267,795 – 16,267,897 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 67.93 |
| Shannon entropy | 0.59476 |
| G+C content | 0.35949 |
| Mean single sequence MFE | -27.40 |
| Consensus MFE | -12.00 |
| Energy contribution | -11.98 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.56 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.960084 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 16267795 102 - 22422827 GCUGUGUUUGCUUGUAAGGAAAUACUU-------CACUUUGCCGAGCAUAAUCCUACAAACGUAUGCAUAUGUACGUGUG------UAUACAUGUUUCUGGGCAAGUUCAGCUUG ((((.....((((((..(((((((...-------.....(((...))).............((((((((((....)))))------))))).)))))))..)))))).))))... ( -31.00, z-score = -2.36, R) >droSim1.chrX_random 4279690 115 - 5698898 GCUGUGUUCGCUUGUAAGGAAAUACUUUAUAAUACACUUUGCCGAGCAUAAUCCUACCAACGUAUGCAUAUGUACGUGUGUAUAUAUGUAUAUGUUUCUGGGCAAGUUCAGCUUG (((((((....((((((((.....)))))))).))))......((((.....((((..(((((((((((((((((....)))))))))))))))))..))))...)))))))... ( -36.90, z-score = -3.55, R) >droSec1.super_17 84793 113 - 1527944 GCUGUGUUUGCUUGUAAGGAAAUACUUUAUAAUACACUUUGCCGAGCAUAAUCCUACCAACGUAUGCAUAUGUACGUGUG--UAUAUGUAUAUGUUUCUGGGCAAGUUCAGCUUG ((((.....((((((..(((((((...((((((((((..(((...)))...........((((((......)))))))))--))).))))..)))))))..)))))).))))... ( -32.30, z-score = -2.09, R) >droEre2.scaffold_4690 7919018 97 + 18748788 GAUGUGUCUGCUUUUAAGGAAAUACUU-------CACUAUGCCGAACAUAAUCCUACAAACAUAUAUGUACA-AUGUAUG----------UAUGUUUCUGGACAAGUUCAGCUUG ((.((((.(.((....)).).)))).)-------).....((.((((....(((.(((.((((((((.....-)))))))----------).)))....)))...)))).))... ( -21.70, z-score = -1.78, R) >droYak2.chrX 10387491 98 - 21770863 GAUGUGUUUGCUUGUAAGGAAAUACUU-------CACUUUGCCGAACAUAAUCCUACAAACAUGUUUGUAUGUACUUUUG----------UAUGUUUCUGGGCAAGUUCAGCUUG ((.((((((.((....)).)))))).)-------)..(((((((((((((....((((.(((....))).))))......----------)))))))...))))))......... ( -22.80, z-score = -1.37, R) >droAna3.scaffold_13186 45483 106 + 49402 UAUGUAUGUAUAUGUACUAACAUACGUACAUAUCUACAAUAUUAAUUCCAGUUCU---GGUAUAUUUUGAGACACUGCUU-----AACAAUAGUUUUUUGUUUGUGUUUAUGUA- (((((((((((.(......).)))))))))))..((((.........(((....)---)).........((((((.....-----(((((.......))))).)))))).))))- ( -19.70, z-score = -0.37, R) >consensus GAUGUGUUUGCUUGUAAGGAAAUACUU_______CACUUUGCCGAGCAUAAUCCUACAAACAUAUGCAUAUGUACGUGUG______U_UAUAUGUUUCUGGGCAAGUUCAGCUUG ((((.....((((((..(((((((...................((......))......((((((......))))))...............)))))))..)))))).))))... (-12.00 = -11.98 + -0.02)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:49:25 2011