
| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,229,432 – 16,229,537 |
| Length | 105 |
| Max. P | 0.658225 |

| Location | 16,229,432 – 16,229,537 |
|---|---|
| Length | 105 |
| Sequences | 8 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 70.65 |
| Shannon entropy | 0.56013 |
| G+C content | 0.48052 |
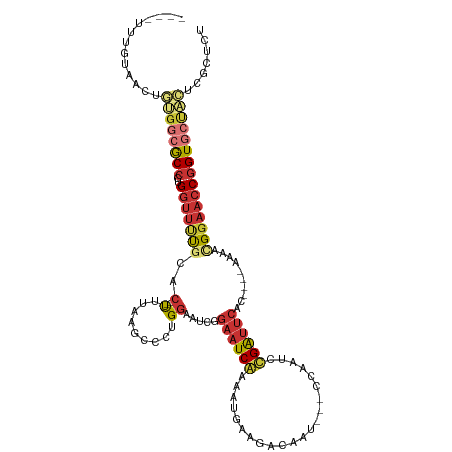
| Mean single sequence MFE | -27.75 |
| Consensus MFE | -14.24 |
| Energy contribution | -15.26 |
| Covariance contribution | 1.02 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.658225 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
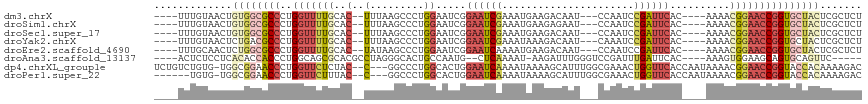
>dm3.chrX 16229432 105 + 22422827 ----UUUGUAACUGUGGCGCCCUGGUUUUGCAC--UUUAAGCCCUGGAAUCGGAAUCGAAAUGAAGACAAU---CCAAUCCGAUUCAC----AAAACGGAACCGGUGCUACUCGCUCU ----.........((((((((..((((((((..--.....))..((((((((((...((...........)---)...)))))))).)----)....))))))))))))))....... ( -28.50, z-score = -1.58, R) >droSim1.chrX 12522201 105 + 17042790 ----UUUGUAACUGUGGCGCCCUGGUUUUGCAC--UUUAAGCCCUGGAAUCGGAAUCGAAAUGAAGAGAAU---CCAAUCCGAUUCAC----AAAACGGAACCGGUGCUACUCGCUCU ----.........((((((((..((((((((..--.....))..((((((((((...((...........)---)...)))))))).)----)....))))))))))))))....... ( -28.50, z-score = -1.34, R) >droSec1.super_17 56434 105 + 1527944 ----UUUGUAACUGUGGCGCCCUGGUUUUGCAC--UUUAAGCCCUGGAAUCGGAAUCGAAAUGAAGACAAU---CCAAUCCGAUUCAC----AAAACGGAACCGGUGCUACUCGCUCU ----.........((((((((..((((((((..--.....))..((((((((((...((...........)---)...)))))))).)----)....))))))))))))))....... ( -28.50, z-score = -1.58, R) >droYak2.chrX 10360326 105 + 21770863 ----UUUGUAACUCUGACGCCCUGGUUUUGCAC--UUUAAGCCCUGGAAUCGGAAUCGAAAUAAAGACAAU---CAAAUCCGAUUCAC----AAAACGGAACCGGUGCUACUCGCUCU ----..............((.((((((((((..--.....))..((((((((((...((...........)---)...)))))))).)----)....)))))))).)).......... ( -22.10, z-score = -0.73, R) >droEre2.scaffold_4690 7889797 105 - 18748788 ----UUUGCAACUCUGGCGCCCUGGUUUUGCAC--UAUAAGCCCUGGAAUCGGAAUCAAAAUGAAGACAAU---CCAAUCCGAUUCAC----AAAACGGAACCGGUGCUACUCGCUCU ----...((.....(((((((..((((((((..--.....))..((((((((((.((.....)).(.....---.)..)))))))).)----)....)))))))))))))...))... ( -25.50, z-score = -1.22, R) >droAna3.scaffold_13137 314318 102 - 1373759 ----ACUCUCCUCACACCACCCUGGCAGCGCACGCCUAGGGCACUGCCAAUG--CUCAAAAU-AAGAUUUGGGUCCGAUUUGAUUCAC----AAAGUGGAAGCAGUGCAGUUC----- ----................((((((.......))).)))(((((((.....--((((((..-....))))))((((.((((.....)----))).)))).))))))).....----- ( -29.50, z-score = -0.82, R) >dp4.chrXL_group1e 9811941 112 + 12523060 UCUGUCUGUG-UGGCGGAACCCUGGUUCUCUAC--C---GGCCCUGGCACUGGAAUCAAAAUAAAAGCAUUUGGCGAAACUGGUUCACCAAUAAAACGGAACCGGUACCACAAAAGAC ...((((.((-(((.(((((....))))).(((--(---((..(((((..................))..((((.(((.....))).)))).....)))..)))))))))))..)))) ( -31.17, z-score = -0.99, R) >droPer1.super_22 1298313 106 - 1688296 ------UGUG-UGGCGGAACCCUGGUUCUUUAC--C---GGCCCUGGCACUGGAAUCAAAAUAAAAGCAUUUGGCGAAACUGGUUCACCAAUAAAACGGAACCGGUACCACAAAAGAC ------..((-(((.(((((....))))).(((--(---((..(((((..................))..((((.(((.....))).)))).....)))..)))))))))))...... ( -28.27, z-score = -0.48, R) >consensus ____UUUGUAACUGUGGCGCCCUGGUUUUGCAC__UUUAAGCCCUGGAAUCGGAAUCAAAAUGAAGACAAU___CCAAUCCGAUUCAC____AAAACGGAACCGGUGCUACUCGCUCU .............((((((((..(((((((...........(....).....((((((......................))))))..........)))))))))))))))....... (-14.24 = -15.26 + 1.02)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:49:21 2011