| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,216,836 – 16,216,916 |
| Length | 80 |
| Max. P | 0.782178 |

| Location | 16,216,836 – 16,216,916 |
|---|---|
| Length | 80 |
| Sequences | 10 |
| Columns | 86 |
| Reading direction | forward |
| Mean pairwise identity | 67.22 |
| Shannon entropy | 0.69242 |
| G+C content | 0.52549 |
| Mean single sequence MFE | -30.42 |
| Consensus MFE | -8.71 |
| Energy contribution | -8.11 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.82 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.782178 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
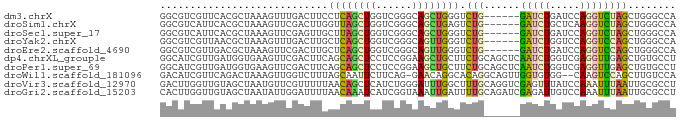
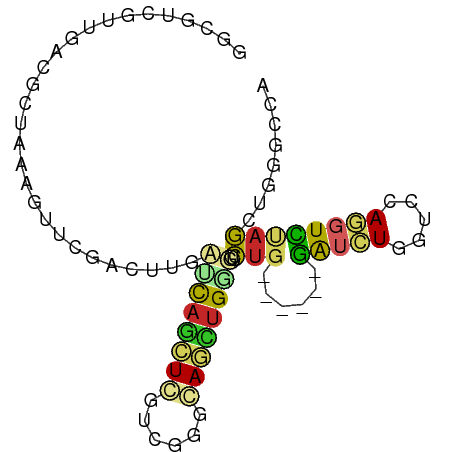
>dm3.chrX 16216836 80 + 22422827 GGCGUCGUUCACGCUAAAGUUUGACUUCCUCAGCUGGUCGGGCAGCUGGGUCUG------GAUCUGAUCCAGGUCUAGCUGGGCCA (((.(((..((.(((.(((.....)))....)))))..))).(((((((..(((------(((...))))))..))))))).))). ( -34.10, z-score = -2.23, R) >droSim1.chrX 12509295 80 + 17042790 GGCGUCAUUCACGCUAAAGUUCGACUUGGUUAGCUGGUCGGGCAGCUGAGUCUG------GAUCUGCUCAAGGUCUAGCUGGGCCA (((((.....)))))...((((((((.(.....).)))))))).(((.((.(((------(((((.....)))))))))).))).. ( -32.00, z-score = -2.11, R) >droSec1.super_17 44005 80 + 1527944 GGCGUCAUUCACGCUAAAGUUCGAGUUGCUUAGCUGGUCGGGCAGCUGGGUCUG------GAUCUGAUCCAGGUCUAGCUGGGCCA ((((((..(((.(((((.((.......))))))))))...)))((((((..(((------(((...))))))..))))))..))). ( -35.70, z-score = -2.98, R) >droYak2.chrX 10347375 80 + 21770863 GGCGUCGUUAACGCUAAAGUUUGACUUGCUCAGCUGGUCGGGCAGUUGGGUCUG------GAUCUGGUCCAGGUCCAGCUGGGCCA (((...............((((((((.((...)).))))))))((((((..(((------(((...))))))..))))))..))). ( -35.90, z-score = -2.88, R) >droEre2.scaffold_4690 7877230 80 - 18748788 GGCGUCGUUGACGCUAAAGUUCGACUUGCUCAGCUGGUCGGGCAGUUGGGUCUG------GAUCUGAUCCAGGUCCAGCUGGGCCA ((((((...))))))...((((((((.((...)).))))))))((((((..(((------(((...))))))..))))))...... ( -39.10, z-score = -3.44, R) >dp4.chrXL_group1e 10377205 86 + 12523060 GGCAUCGUUGAUGGUGAAGUUCGACUUCAGCAGCUCCUCCGGAAGCUGCUUCUGCAGCUCAAUCUGGUCGAGGUUGAGCUGUGCCU (((...........(((((.....)))))((((((((...)).))))))....((((((((((((.....))))))))))))))). ( -37.70, z-score = -2.71, R) >droPer1.super_69 199106 86 - 344178 GGCAUCGUUGAUGGUGAAGUUCGACUUCAGCAGCUCCUCCGGAAGCUGCUUCUGCAGCUCAAUCUGGUCGAGGUUGAGCUGUGCCU (((...........(((((.....)))))((((((((...)).))))))....((((((((((((.....))))))))))))))). ( -37.70, z-score = -2.71, R) >droWil1.scaffold_181096 3242768 83 - 12416693 GACAUCGUUCAGACUAAAGUUGGUCUUUAGCAAUUCUUCAG-GAACAGGCACAGGCAGUUGGUGUGG--CAAGUCCAGCUUGUCCA .((((((((((((((((((.....))))))....)))....-))))..((....))....)))))((--((((.....)))))).. ( -19.30, z-score = 0.35, R) >droVir3.scaffold_12970 2579793 86 - 11907090 GACUUGGUUGUAGCUAAUGUUCGUUUUUAACAGCUCAUCUGGGAUUUGGCUUUGCAGGUCGAGUUUAUCCAAAUUUAAUUGCGCCU ..((..(.((.((((..((........))..)))))).)..))....(((...((((...((((((....))))))..))))))). ( -16.20, z-score = 0.65, R) >droGri2.scaffold_15203 2065529 86 + 11997470 CACUUGGUUGUAGCUAAUAUUGGAUUUUAACAAAUCAUCGGUAAAUUGAUUUUGCAGAUCGAGAUUGUCCAAAUUUAAUUGCGCCU .....(((.((((.(((..((((((..(......(((((.(((((.....))))).))).)).)..))))))..))).))))))). ( -16.50, z-score = -0.68, R) >consensus GGCGUCGUUGACGCUAAAGUUCGACUUGAUCAGCUCGUCGGGCAGCUGGGUCUG______GAUCUGGUCCAGGUCUAGCUGGGCCA .................((((.(((((.((((((((......))))))))....................))))).))))...... ( -8.71 = -8.11 + -0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:49:19 2011