
| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,200,248 – 16,200,394 |
| Length | 146 |
| Max. P | 0.951019 |

| Location | 16,200,248 – 16,200,363 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 71.87 |
| Shannon entropy | 0.51695 |
| G+C content | 0.51432 |
| Mean single sequence MFE | -22.61 |
| Consensus MFE | -14.77 |
| Energy contribution | -15.27 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.926884 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
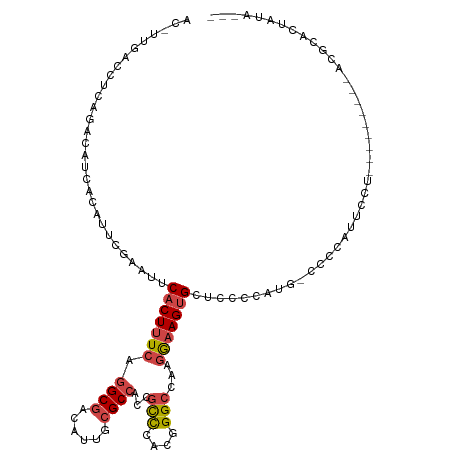
>dm3.chrX 16200248 115 - 22422827 AC-UUGACCUCAGACAUCACAUUCGAAUUCACUUUCAGGCGACAUUGCGCCACCGCCCACGGGCCAAGGAAGUGCUCCCCCAUUCCCCAUUCCUACGCAAUGCACGCUCUCUAUCC ..-........(((..........(((......))).((((.(((((((.....(((....)))..(((((..................))))).)))))))..)))).))).... ( -23.77, z-score = -1.05, R) >droSim1.chrX_random 4218805 106 - 5698898 AC-UUGACCUCAGACAUCACAUUCGAAUUCACUUUCAGGCGACAUUGCGCCACCGCCCACGGGCCAAGGAAGUGCUCCCCAUGGCCCCAUUCCU---------ACCCUCUCUAUCC ..-........(((..........(((......))).((((......)))).........((((((.((.........)).)))))).......---------......))).... ( -21.80, z-score = -0.37, R) >droSec1.super_17 26924 115 - 1527944 AC-UUGACCUCAGACAUCACAUUCGAAUUCACUUUCAGGCGACAUUGCGCCACCGCCCACGGGCCAAGGAAGUGCUCCCCAUGCCCCCAUUCCUACGCAAUGCACGCCCUAUAUCC ..-.....................(((......))).((((.(((((((.....(((....)))..(((((..((.......)).....))))).)))))))..))))........ ( -27.80, z-score = -1.99, R) >droYak2.chrX_random 42905 98 - 1802292 AC-UUGACCUCAGACAUCACAUUUUAAUUCACUUUCAGGCGACAUUGCGCCACCGCCCACGGGCCAAGAAAGUGCUCCCCAUG--UCUACGCA----------AUGCAUUG----- ..-........((((((............(((((((.((((......))))...(((....)))...)))))))......)))--))).....----------........----- ( -23.47, z-score = -1.94, R) >droEre2.scaffold_4690 7861004 101 + 18748788 ACGUCGACCUCAGACAUCACAUUUUAAUUCACUUUCAGGCGACAUUGCGCCACCGCCCACGGGCUAAGAAAGUGCUCCCCACU--CCUAUGCA----------AUGUACAAUG--- ..(((.......)))...(((((.......((((((.((((......))))...(((....)))...))))))((........--.....)))----------))))......--- ( -24.02, z-score = -2.27, R) >droAna3.scaffold_13137 1093510 94 - 1373759 AC-UUAACCUUGGACAUAACAAAUAAAUUCACUUUCAAGCGACAUUG-GCC-CUAUUUAUAGGACAAUGCAGGGAUUGCAAUGCGUACUUUCCAUCU------------------- ..-.....((((((...................)))))).(.(((((-..(-(((....)))).)))))).((((.(((.....)))..))))....------------------- ( -14.81, z-score = -0.10, R) >consensus AC_UUGACCUCAGACAUCACAUUCGAAUUCACUUUCAGGCGACAUUGCGCCACCGCCCACGGGCCAAGGAAGUGCUCCCCAUG_CCCCAUUCCU_________ACGCACUAUA___ .............................(((((((.((((......))))...(((....)))...))))))).......................................... (-14.77 = -15.27 + 0.50)

| Location | 16,200,288 – 16,200,394 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 86.31 |
| Shannon entropy | 0.26015 |
| G+C content | 0.45689 |
| Mean single sequence MFE | -27.55 |
| Consensus MFE | -19.24 |
| Energy contribution | -19.97 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.951019 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
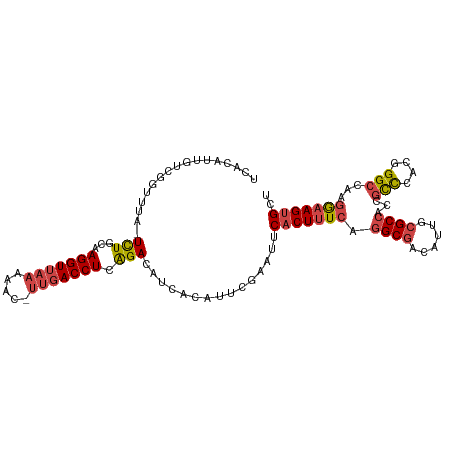
>dm3.chrX 16200288 106 - 22422827 UCACAUUGUCGGUUUA-UCUGCAAGGUUAAAAAC-UUGACCUCAGACAUCACAUUCGAAUUCACUUUCAGGCGACAUUGCGCCACCGCCCACGGGCCAAGGAAGUGCU ......(((.(((...-((((..(((((((....-))))))))))).))))))........(((((((.((((......))))...(((....)))...))))))).. ( -30.60, z-score = -2.23, R) >droSim1.chrX_random 4218836 106 - 5698898 UCACAUUGUCUUUUUA-UCUGCAAGGUUAAAAAC-UUGACCUCAGACAUCACAUUCGAAUUCACUUUCAGGCGACAUUGCGCCACCGCCCACGGGCCAAGGAAGUGCU ................-((((..(((((((....-)))))))))))...............(((((((.((((......))))...(((....)))...))))))).. ( -29.20, z-score = -2.52, R) >droSec1.super_17 26964 106 - 1527944 UCACAUUGUCGUUUUA-UCUGCAAGGUUAAAAAC-UUGACCUCAGACAUCACAUUCGAAUUCACUUUCAGGCGACAUUGCGCCACCGCCCACGGGCCAAGGAAGUGCU ......((((((((..-((((..(((((((....-)))))))))))..........(((......)))))))))))..((((..(((((....)))...))..)))). ( -29.90, z-score = -2.40, R) >droYak2.chrX_random 42928 106 - 1802292 UCACACUGUCGGUUUA-UCUUCAAGGUUACCAAC-UUGACCUCAGACAUCACAUUUUAAUUCACUUUCAGGCGACAUUGCGCCACCGCCCACGGGCCAAGAAAGUGCU ......((((((....-...)).((((((.....-.))))))..)))).............(((((((.((((......))))...(((....)))...))))))).. ( -27.90, z-score = -2.75, R) >droEre2.scaffold_4690 7861029 107 + 18748788 UCACAUUGUCGGUUUA-UCUUCAAGGUUAACAACGUCGACCUCAGACAUCACAUUUUAAUUCACUUUCAGGCGACAUUGCGCCACCGCCCACGGGCUAAGAAAGUGCU ......((((((....-...)).((((..((...))..))))..)))).............(((((((.((((......))))...(((....)))...))))))).. ( -28.30, z-score = -2.86, R) >droAna3.scaffold_13137 1093531 103 - 1373759 UCACAUU--CGCUUUAUUUUUCAAGGUCAAUAAC-UUAACCUUGGACAUAACAAAUAAAUUCACUUUCAAGCGACAUUG-GCC-CUAUUUAUAGGACAAUGCAGGGAU ......(--((((((((((((((((((.((....-)).))))))))......)))))...........))))))(((((-..(-(((....)))).)))))....... ( -19.40, z-score = -2.35, R) >consensus UCACAUUGUCGGUUUA_UCUGCAAGGUUAAAAAC_UUGACCUCAGACAUCACAUUCGAAUUCACUUUCAGGCGACAUUGCGCCACCGCCCACGGGCCAAGGAAGUGCU .................(((...(((((((.....))))))).)))...............(((((((.((((......))))...(((....)))...))))))).. (-19.24 = -19.97 + 0.72)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:49:17 2011