
| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,192,795 – 16,192,914 |
| Length | 119 |
| Max. P | 0.586405 |

| Location | 16,192,795 – 16,192,914 |
|---|---|
| Length | 119 |
| Sequences | 10 |
| Columns | 137 |
| Reading direction | forward |
| Mean pairwise identity | 62.62 |
| Shannon entropy | 0.75720 |
| G+C content | 0.44417 |
| Mean single sequence MFE | -31.10 |
| Consensus MFE | -10.00 |
| Energy contribution | -9.59 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.586405 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
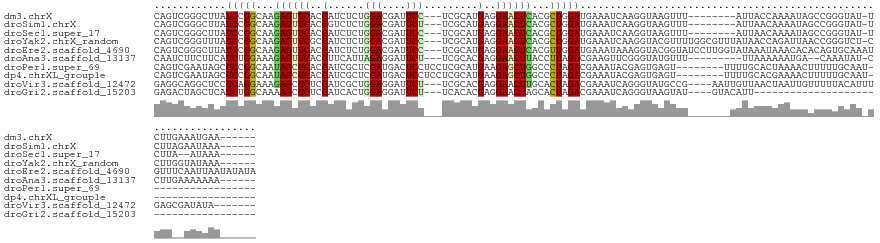
>dm3.chrX 16192795 119 + 22422827 CAGUCGGGCUUAUCCGGCAAGAGUUCACCAUCUCUGGACGAUUCC---UCGCAUGAGGAACUCACGCUGGAUGAAAUCAAGGUAAGUUU--------AUUACCAAAAUAGCCGGGUAU-UCUUGAAAUGAA------ ...(((((..((((((((..((.((((.....((..(..((((((---((....)))))).))...)..)))))).))..(((((....--------.)))))......)))))))).-.)))))......------ ( -39.80, z-score = -3.13, R) >droSim1.chrX 12495864 119 + 17042790 CAGUCGGGCUUAUCCGGCAAGAGUUCACCGUCUCUGGACGAUUCU---UCGCAUGAGGAACUCACGCUGGAUGAAAUCAAGGUAAGUUU--------AUUAACAAAAUAGCCGGGUAU-UCUUAGAAUAAA------ ...((.((((((((((((..((((((..((((....)))).(((.---......)))))))))..)))))))))...........(((.--------...)))......))).))(((-((...)))))..------ ( -33.60, z-score = -1.92, R) >droSec1.super_17 19571 117 + 1527944 CAGUCGGGCUUAUCCGGCAAGAGUUCACCAUCUCUGGACGAUUCC---UCGCAUGAGGAACUCACGCUGGAUGAAAUCAAGGUAAGUUU--------AUUAACAAAAUAGCCGGGUAU-UCUUA--AUAAA------ ...((.((((((((((((..((((((..(((....(((....)))---....)))..))))))..)))))))))...........(((.--------...)))......))).))...-.....--.....------ ( -33.90, z-score = -2.39, R) >droYak2.chrX_random 35860 127 + 1802292 CAGUCGGGUUUAUCCGGCAAGAGUUCGCCAUCUCUGGACGAUUCC---UCGCAUGAGGAACUCACGCUGGAUGAAAUCAAGGUACGUUUUGGCGUUUAUAACCAGAUUAACCGGGUCU-CCUUGGUAUAAA------ .(((((..((((((((((..((((((((((....))).))).(((---((....)))))))))..))))))))))..)..(((((((....)))).....))).)))).((((((...-.)))))).....------ ( -42.50, z-score = -2.33, R) >droEre2.scaffold_4690 7853783 134 - 18748788 CAGUCGGGCUUAUCCGGCAAGAGUUCACCAUCUCUGGACGAUUCC---UCGCAUGAGGAACUCACGUUGGAUGAAAUAAAGGUACGGUAUCCUUGGUAUAAAUAAACACACAGUGCAAAUGUUUCAAUUAAUAUAUA ..((((((....))))))..((((((..(((....(((....)))---....)))..))))))..(((((.(((((((...((((.((((.....)))).............))))...))))))).)))))..... ( -32.94, z-score = -1.15, R) >droAna3.scaffold_13137 201074 116 - 1373759 CAAUCUUCUUCAUCUGGAAAGAGUUCACCUUCAUUAGAGGAUUCU---UCGCACGAGGAACUUACCUUAGACGAAGUUCGGGUAUGUUU---------UUAAAAAAUGA--CAAAUAU-CCUUGAAAAAAA------ .......((((.(((((.(((......((((.....)))).((((---((....))))))))).))..))).))))((((((.((((((---------(((.....)))--.))))))-.)))))).....------ ( -22.50, z-score = -0.05, R) >droPer1.super_69 115866 111 + 344178 CAGUCGAAUAGCUCCGGCAAUAGCUCACCAUCGCUCGAUGACUCCUCCUCGCAUGAAGAGCUGGCCCUAGACGAAAUACGAGUGAGU--------UUUUGCACUAAAACUUUUUGCAAU------------------ ..(.((((.((.....((((.(((((((((((....))))...((..(((.......)))..)).................))))))--------).)))).......)).)))))...------------------ ( -24.00, z-score = 0.20, R) >dp4.chrXL_group1e 2119123 111 - 12523060 CAGUCGAAUAGCUCCGGCAAUAGCUCACCAUCGCUCGAUGACUCCUCCUCGCAUGAAGAGCUGGCCCUAGACGAAAUACGAGUGAGU--------UUUUGCACGAAAACUUUUUGCAAU------------------ ..(.((((.((...((((((.(((((((((((....))))...((..(((.......)))..)).................))))))--------).)))).))....)).)))))...------------------ ( -25.80, z-score = -0.07, R) >droVir3.scaffold_12472 507056 123 - 763072 GAGGCAGGCUCCUCAGGAAAGAGCUCUCCAUCGCUGGAGGAUUCU---UCGCACGAGGAACUUGCACUAGACGAAAUCAGGGUAUGCCG----AAUUGUUAACUAAUUGUUUUUACAUUUGAGCGAUAUA------- .(((((((.(((((..(.(((((..(((((....)))))..))))---)..)..))))).))))).)).............((((((((----(((.((.(((.....)))...))))))).)).)))).------- ( -35.60, z-score = -1.27, R) >droGri2.scaffold_15203 4728748 93 + 11997470 GAGACUAGCUCAUCUGGCAAAAGCUCUCCAUCACUGGAGGAUUCU---UCACACGAGGAACUAGCACUAGACGAAAUCAGGGUAAGUAU----GUACAUU------------------------------------- ...(((.((((.(((((.......((((((....)))))).((((---((....))))))......))))).(....).)))).)))..----.......------------------------------------- ( -20.40, z-score = -0.26, R) >consensus CAGUCGGGCUUAUCCGGCAAGAGUUCACCAUCUCUGGACGAUUCC___UCGCAUGAGGAACUCACGCUAGACGAAAUCAAGGUAAGUUU_______UAUUAACAAAAUAGCCGGGUAU_UCUU__AAUAAA______ ............(((((...((((((..(......(((....))).........)..))))))...))))).................................................................. (-10.00 = -9.59 + -0.41)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:49:15 2011