| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,183,154 – 16,183,246 |
| Length | 92 |
| Max. P | 0.585921 |

| Location | 16,183,154 – 16,183,246 |
|---|---|
| Length | 92 |
| Sequences | 12 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 57.46 |
| Shannon entropy | 0.91369 |
| G+C content | 0.44361 |
| Mean single sequence MFE | -22.65 |
| Consensus MFE | -8.90 |
| Energy contribution | -8.76 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.60 |
| Mean z-score | -0.46 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.585921 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
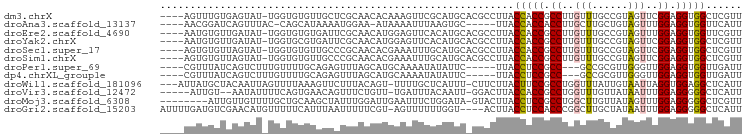
>dm3.chrX 16183154 92 - 22422827 ----AGUUUGUGAGUAU-UGGUGUGUUGCUCGCAACACAAAGUUCGCAUGCACGCCUUACCACCGCCUUGUUUGCCGUAGUUCGGAGGUGGCUCGUU ----.......(((...-.((((((((((..((........))..))).)))))))......((((((......(((.....))))))))))))... ( -26.40, z-score = 0.15, R) >droAna3.scaffold_13137 164574 86 + 1373759 ----AACGGAUCAGUUUAC-CAGCAUAAAAUGGAA-AUAAAAUUUAAGUGC-----UUACCACCACCUUGCUUGCUGUAGUUUGGAGGUGGUUCAUU ----.........((((.(-((........)))))-))........((((.-----..((((((.((..(((......)))..)).)))))).)))) ( -20.10, z-score = -0.81, R) >droEre2.scaffold_4690 7844314 92 + 18748788 ----AAUGUGUUGAUAU-UGGUGUGUGAUUCGCAACAUGGAGUUCACAUGCACGCCUUACCACCGCCUUGUUUGCCGUAGUUUGGAGGUGGUUCGUU ----.............-..(((((((((((........))).))))))))(((....((((((.((................)).)))))).))). ( -23.39, z-score = 0.54, R) >droYak2.chrX 10328838 92 - 21770863 ----AAUGUGUUGAUAU-UGGUGCGUGAUUCGCAACAUGGAGUUCACAUGCACGCCUUACCACCGCCUUGUUUGCCGUAGUUCGGAGGUGGCUCGUU ----.(((((.......-((.((((.....)))).)).......)))))((.((((((((.((.((.......)).)).))...))))))))..... ( -22.34, z-score = 1.44, R) >droSec1.super_17 10072 92 - 1527944 ----AGUGUGUUAGUAU-UGGUGUGUUGCCCGCAACACGAAAUUUGCAUGCACGCCUUACCACCGCCUUGUUUGCCGUAGUUCGGAGGUGGCUCGUU ----.((((((..(((.-...(((((((....))))))).....)))..)))))).......((((((......(((.....)))))))))...... ( -28.40, z-score = -0.55, R) >droSim1.chrX 12485854 92 - 17042790 ----AGUGUGUUAGUAU-UGGUGUGUUGCCCGCAACACGAAAUUUGCAUGCACGCCUUACCACCGCCUUGUUUGCCGUAGUUCGGAGGUGGCUCGUU ----.((((((..(((.-...(((((((....))))))).....)))..)))))).......((((((......(((.....)))))))))...... ( -28.40, z-score = -0.55, R) >droPer1.super_69 103751 85 - 344178 ----CGUUUAUCAGUCUUUGUUUUGCAGAGUUUAGCAUGCAAAAUAUAUUC-----UUACCUCCGCC---GCCGCGUUGGGUUGGAGGUGGUUGAUU ----.....((((..(..(((((((((..(.....).))))))))).....-----..(((((((..---(((......)))))))))))..)))). ( -25.20, z-score = -2.04, R) >dp4.chrXL_group1e 2107113 85 + 12523060 ----CGUUUAUCAGUCUUUGUUUUGCAGAGUUUAGCAUGCAAAAUAUAUUC-----UUACCUCCGCC---GCCGCGUUGGGUUGGAGGUGGUUGAUU ----.....((((..(..(((((((((..(.....).))))))))).....-----..(((((((..---(((......)))))))))))..)))). ( -25.20, z-score = -2.04, R) >droWil1.scaffold_181096 6535105 92 - 12416693 ---AUUAUGCUACAAUUAGUUUUAAAGUUCUUUACAGU-UUUUGCUCAUUU-CUUCUUACUUCCGCCUGGUUUAUUGUAAUUAGGUGGAGGCUCAUU ---.(((.((((....))))..))).............-............-.......(((((((((((((......)))))))))))))...... ( -18.40, z-score = -1.91, R) >droVir3.scaffold_12472 495399 88 + 763072 -----AUUGU--AAUAUUUUCAGUGAACAGUUUCUGUU-UGAUUUACAAUU-GGACUUACCACCGCCUGGUUUGUUAUAAUUUGGAGGGGGCUCAUU -----(((((--((.....(((...(((((...)))))-))).))))))).-.......((.((.((..(((......)))..)).))))....... ( -14.80, z-score = 0.88, R) >droMoj3.scaffold_6308 3213250 88 - 3356042 --------AUUGUUGUUUUGCUGCAAGCUAUUUGGAUUGAAUUUCUGGAUA-GUACUUACCUCCGCCUGGCUUGUUAUAGUUUGGAGGGGGCUCGUU --------.((((.(.....).))))((((((..((.......))..))))-)).....(((((.((..(((......)))..)).)))))...... ( -25.80, z-score = -1.73, R) >droGri2.scaffold_15203 4715558 92 - 11997470 AUUUUGAUGUCGAACAUGUUUUUCAUUUAAUUUUUCGU-AGUUUUUUGGU----ACUUACCUCCACCCGGCUUGCUAUAAUUUGGAGGGGGCUCAUU ....(((.(((.........................((-(.(.....).)----))...((((((...((....))......)))))).)))))).. ( -13.40, z-score = 1.17, R) >consensus ____AGUUUGUUAGUAU_UGGUGUGUAGAAUGUAACAUGAAAUUUACAUGC_CGCCUUACCACCGCCUUGUUUGCCGUAGUUUGGAGGUGGCUCAUU ...........................................................(((((.((..(((......)))..)).)))))...... ( -8.90 = -8.76 + -0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:49:14 2011