| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,176,302 – 16,176,417 |
| Length | 115 |
| Max. P | 0.577125 |

| Location | 16,176,302 – 16,176,417 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 70.05 |
| Shannon entropy | 0.47964 |
| G+C content | 0.43708 |
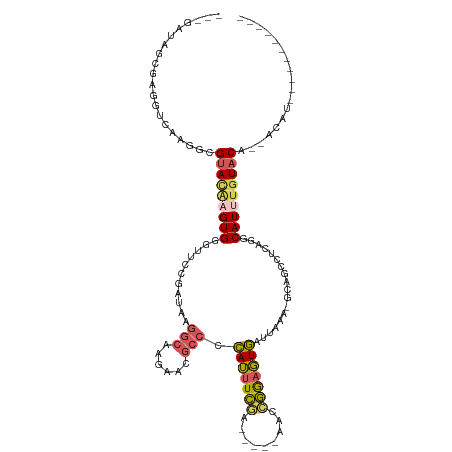
| Mean single sequence MFE | -23.00 |
| Consensus MFE | -14.62 |
| Energy contribution | -15.02 |
| Covariance contribution | 0.40 |
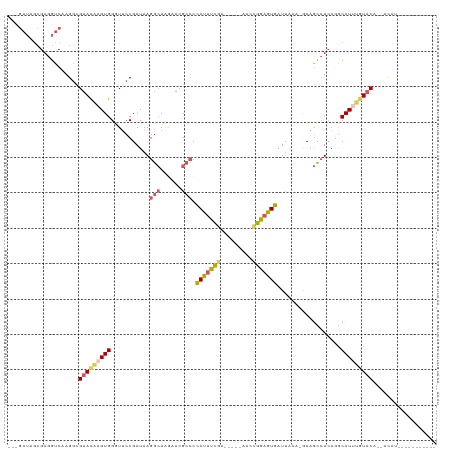
| Combinations/Pair | 1.37 |
| Mean z-score | -0.91 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.577125 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
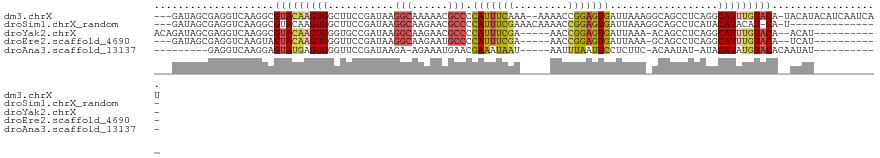
>dm3.chrX 16176302 115 - 22422827 ---GAUAGCGAGGUCAAGGCGUACAAGUGGCUUCCGAUAAGGCAAAAACGCCCCAUUUCAAA--AAAACCGGAGUGAUUAAAGGCAGCCUCAGGCAUUUGUACA-UACAUACAUCAAUCAU ---(((.((.........))((((((((((((.((.....(((......))).((((((...--......))))))......)).))))......)))))))).-.......)))...... ( -24.90, z-score = -0.73, R) >droSim1.chrX_random 4191642 101 - 5698898 ---GAUAGCGAGGUCAAGGCGUACAAGUGGCUUCCGAUAAGGCAAGAACGCCCCAUUUCGAAACAAAACCGGAGUGAUUAAAGGCAGCCUCAUACAUACAU-CA-U--------------- ---......(((((...(((((.(...((.(((.....))).)).).))))).(((((((.........)))))))..........)))))..........-..-.--------------- ( -23.90, z-score = -1.18, R) >droYak2.chrX 10322127 102 - 21770863 ACAGAUAGCGAGGUCAAGGCGUACAAGUGGGUGCCGAUAAGGCAAGAACGCCCCAUUUCGA-----AACCGGAGUGAUUAAA-ACAGCCUCAGGCAUUUGUACA--ACAU----------- ((((((.(((((((...(((((.(....)..((((.....))))...))))).(((((((.-----...)))))))......-...)))))..))))))))...--....----------- ( -30.70, z-score = -2.46, R) >droEre2.scaffold_4690 7837546 99 + 18748788 ---GAUAGCGAGGUCAAGUAGUACAAGUGGGUUCCGAUAAGGCAAGAAUGCCCCAUUUCGA-----AACCGGAGUGAUUAAA-GCAGCCUCAGGCAUUUGUACA--UCAU----------- ---(((......))).....(((((((((((((.(.....((((....)))).(((((((.-----...)))))))......-).)))).....))))))))).--....----------- ( -25.10, z-score = -1.41, R) >droAna3.scaffold_13137 140360 93 + 1373759 ---------GAGGUCAAGGAGUAUGAGUGGGUUCCGAUAAGA-AGAAAUGAACGAAAUAAU-----AAUUUAAUUCCUCUUC-ACAAUAU-AUACAUAUGUACACAAUAU----------- ---------...........(((((.((((....).....((-(((...(((..((((...-----.))))..))).)))))-.......-...))).))))).......----------- ( -10.40, z-score = 1.22, R) >consensus ___GAUAGCGAGGUCAAGGCGUACAAGUGGGUUCCGAUAAGGCAAGAACGCCCCAUUUCGA_____AACCGGAGUGAUUAAA_GCAGCCUCAGGCAUUUGUACA__ACAU___________ ....................(((((((((...........(((......))).(((((((.........)))))))..................))))))))).................. (-14.62 = -15.02 + 0.40)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:49:13 2011