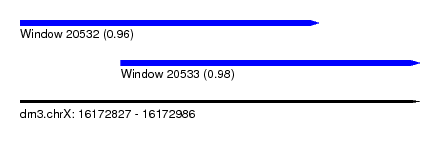
| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,172,827 – 16,172,986 |
| Length | 159 |
| Max. P | 0.983602 |

| Location | 16,172,827 – 16,172,946 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.83 |
| Shannon entropy | 0.14273 |
| G+C content | 0.58814 |
| Mean single sequence MFE | -50.62 |
| Consensus MFE | -44.18 |
| Energy contribution | -45.62 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.961239 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
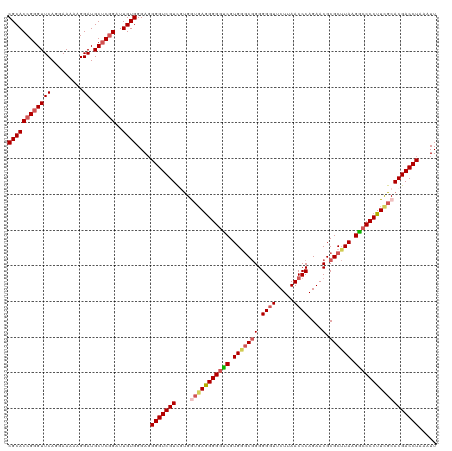
>dm3.chrX 16172827 119 + 22422827 CGCACCGGGUUCAGGAAAAAGGAUGCUCGGAUUGUGGGAUGGGUCGUUGUGGUAAGGGGGUCGUGGGCGGGGG-UUCUUAACCCAGAACGCGCUUAAGGCCCCUUCGCACGACCUAAAUU ((((((((((((.........)).))))))..))))...((((((((.((((...(((((((..(.(((.(((-(.....))))....))).)....))))))))))))))))))).... ( -50.40, z-score = -2.44, R) >droSim1.chrX_random 4188149 120 + 5698898 CGCACCGGGUUCAGGAAAAAGGAUGCUCGGAUUGUGGGAGGGGUCGUCACGGUGAGGGGGUCGUGGGCGGGGGGUUCUUAACCCAGAACGCGCUUAAGGCCCCUUCGCACGACCUAAAUU ((((((((((((.........)).))))))..))))....(((((((....(((((((((((.(((((((.((((.....))))....).)))))).))))))))))))))))))..... ( -54.20, z-score = -2.85, R) >droSec1.super_8489 448 120 + 1584 CGCACCGGGUUCAGGAAAAAGGAUGCUCGGAUUGUGGGAGGGGUCGUCAUGGUGAGGGGGUCGUGGGCGGGGGGUUCUUAACCCAGAACGCGCUUAAGGCCCCUUCGCACGACCUAAAUU ((((((((((((.........)).))))))..))))....(((((((....(((((((((((.(((((((.((((.....))))....).)))))).))))))))))))))))))..... ( -54.20, z-score = -3.08, R) >droYak2.chrX 10316886 118 + 21770863 CGCACCGGGUUCA--AAAAAGGAUGCACGGUUUGUGGGAGGGGUCGUCGUGAGGAAGGGGCCGUGGACAGGGGGUUCUUAACCCAGAACGCGCGUAAGGCCCCUUUGCACGACCUAAAUU (((((((.((((.--.....))))...))))..)))....((((((((....)(((((((((....((.(.(.(((((......))))).).)))..)))))))))..)))))))..... ( -45.80, z-score = -1.28, R) >droEre2.scaffold_4690 7833936 118 - 18748788 CGCACCGGGUUCAG-AAAAAGGAUGCUCCGUUUGUGGGAGGGGUCGUCGUGAUGAGGGG-UCGUGAGCGGGGGGUUCUUAACCCAGAACGCGCUUAAGACCCCUUUGCACGACCUAAAUU ((((.(((((((..-......)).)).)))..))))....(((((((.((...((((((-((.(((((((.((((.....))))....).)))))).)))))))).)))))))))..... ( -48.50, z-score = -2.16, R) >consensus CGCACCGGGUUCAGGAAAAAGGAUGCUCGGAUUGUGGGAGGGGUCGUCGUGGUGAGGGGGUCGUGGGCGGGGGGUUCUUAACCCAGAACGCGCUUAAGGCCCCUUCGCACGACCUAAAUU ((((((((((((.........)).))))))..))))....(((((((....(((((((((((.(((((((.((((.....))))....).)))))).))))))))))))))))))..... (-44.18 = -45.62 + 1.44)


| Location | 16,172,867 – 16,172,986 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.83 |
| Shannon entropy | 0.19314 |
| G+C content | 0.55515 |
| Mean single sequence MFE | -50.62 |
| Consensus MFE | -43.10 |
| Energy contribution | -44.70 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.983602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
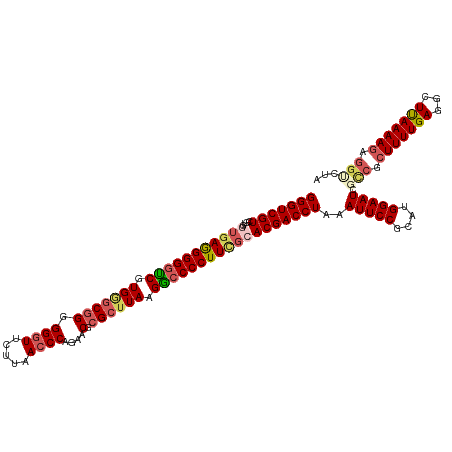
>dm3.chrX 16172867 119 + 22422827 GGGUCGUUGUGGUAAGGGGGUCGUGGGCGGGGG-UUCUUAACCCAGAACGCGCUUAAGGCCCCUUCGCACGACCUAAAUUCCGCAUGGAAUCGCCGCUUUUGAGGCUUAAAAGAGGUCUA (((((((.((((...(((((((..(.(((.(((-(.....))))....))).)....))))))))))))))))))..(((((....))))).(((.(((((((...))))))).)))... ( -48.80, z-score = -1.82, R) >droSim1.chrX_random 4188189 120 + 5698898 GGGUCGUCACGGUGAGGGGGUCGUGGGCGGGGGGUUCUUAACCCAGAACGCGCUUAAGGCCCCUUCGCACGACCUAAAUUCCGCAUGGAAUCGCCGCUUUUGAGGCUUAAAAGAGGCCUU (((((((....(((((((((((.(((((((.((((.....))))....).)))))).))))))))))))))))))..(((((....))))).(((.(((((((...))))))).)))... ( -55.70, z-score = -2.87, R) >droSec1.super_8489 488 120 + 1584 GGGUCGUCAUGGUGAGGGGGUCGUGGGCGGGGGGUUCUUAACCCAGAACGCGCUUAAGGCCCCUUCGCACGACCUAAAUUCCGCAUGGAAUCGCCGCUUUUGAGGCUUAAAAGAGACCUU ((((((((...(((((((((((.(((((((.((((.....))))....).)))))).)))))))))))..)))....(((((....))))).(((........)))........))))). ( -52.00, z-score = -2.36, R) >droYak2.chrX 10316924 120 + 21770863 GGGUCGUCGUGAGGAAGGGGCCGUGGACAGGGGGUUCUUAACCCAGAACGCGCGUAAGGCCCCUUUGCACGACCUAAAUUCCGAAUGGAAUCUUCGAUUUUGAAACUCAAAAGACGUAUA ..(((((((((..(((((((((....((.(.(.(((((......))))).).)))..))))))))).))))))....(((((....)))))......(((((.....))))))))..... ( -44.90, z-score = -2.31, R) >droEre2.scaffold_4690 7833975 119 - 18748788 GGGUCGUCGUGAUGAGGGG-UCGUGAGCGGGGGGUUCUUAACCCAGAACGCGCUUAAGACCCCUUUGCACGACCUAAAUUCCGAUUGGAAUCUCCGCUUUUGAGACUCAAAAGAGGUCUA (((((((.((...((((((-((.(((((((.((((.....))))....).)))))).)))))))).)))))))))..(((((....)))))..((.((((((.....)))))).)).... ( -51.70, z-score = -3.65, R) >consensus GGGUCGUCGUGGUGAGGGGGUCGUGGGCGGGGGGUUCUUAACCCAGAACGCGCUUAAGGCCCCUUCGCACGACCUAAAUUCCGCAUGGAAUCGCCGCUUUUGAGGCUUAAAAGAGGUCUA (((((((....(((((((((((.(((((((.((((.....))))....).)))))).))))))))))))))))))..(((((....))))).(((.(((((((...))))))).)))... (-43.10 = -44.70 + 1.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:49:12 2011