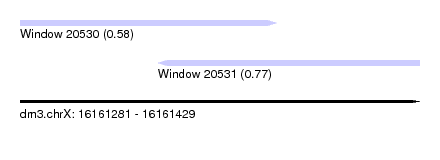
| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,161,281 – 16,161,429 |
| Length | 148 |
| Max. P | 0.772594 |

| Location | 16,161,281 – 16,161,376 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 86.71 |
| Shannon entropy | 0.25362 |
| G+C content | 0.43836 |
| Mean single sequence MFE | -28.93 |
| Consensus MFE | -18.91 |
| Energy contribution | -19.72 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.582860 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
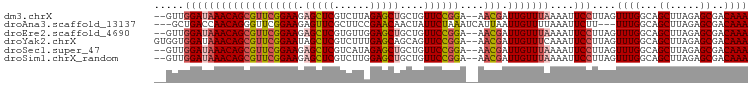
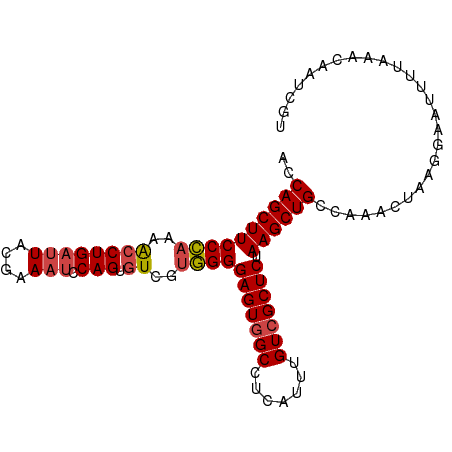
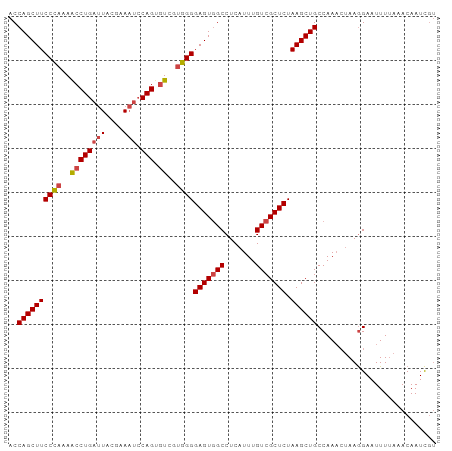
>dm3.chrX 16161281 95 + 22422827 --GUUGGAUAAACAGCGUUCGGAAGAGCUCGUCUUAGAGCUGCUGUUCCGGA--AACGAUUGUUUAAAAUUCCUUAGUUUGGCAGCUUAGAGCGACAAA --((((......))))((((....))))(((.(((.((((((((....((..--..))........((((......)))))))))))).)))))).... ( -31.50, z-score = -2.42, R) >droAna3.scaffold_13137 108089 93 - 1373759 ---GCUGACCAACAGGGUUCGGAAGAGUUCGCUUCCGAACAACUAUUCUAAAUCAUUAAUUGUUUUAAAUUCUU---UUUGGCAGCUUAGAGCGACAAA ---((((.((((.((((((((((((......)))))))))........((((.((.....)).))))....)))---.))))))))............. ( -26.80, z-score = -3.34, R) >droEre2.scaffold_4690 7822807 95 - 18748788 --GUUGGAUAAACAGCGUUCGGAAGAGCUCGUGUUGGAGCUGCUGUUCCGGA--AACGAUUGUUUAAAAUUCCUUAGUUUGGCAGCUUAGAGCGACAAA --((((......))))((((....))))((((....((((((((....((..--..))........((((......))))))))))))...)))).... ( -30.80, z-score = -2.10, R) >droYak2.chrX 10304897 97 + 21770863 GUGGUGGAUAAACAGCGUUCGGAAUAGCUCGUCUUUGAGCAGCAGUUCCGGA--AACGAUUGUUUCAAAUUCCUUAGUUUGGCAGCUUAGAGCGACAAA ((.((((((((((((((((((((((.(((((....)))))....))))))).--.))).))))))...))))((.(((......))).)).)).))... ( -24.20, z-score = -0.05, R) >droSec1.super_47 209505 95 + 216421 --GUUGGAUAAACAGCGUUCGGAAGAGCUCGUCAUAGAGCUGCUGUUCCGGA--AACGAUUGUUUAAAAUUCCUUAGUUUGGCAGCUUAGAGCGACAAA --((((......))))((((....))))((((..(.((((((((....((..--..))........((((......)))))))))))))..)))).... ( -28.80, z-score = -1.80, R) >droSim1.chrX_random 4204541 95 + 5698898 --GUUGGAUAAACAGCGUUCGGAAGAGCUCGUCUUGGAGCUGCUGUUCCGGA--AACGAUUGUUUAAAAUUCCUUAGUUUGGCAGCUUAGAGCGACAAA --((((......))))((((....))))(((.(((.((((((((....((..--..))........((((......)))))))))))).)))))).... ( -31.50, z-score = -2.22, R) >consensus __GUUGGAUAAACAGCGUUCGGAAGAGCUCGUCUUAGAGCUGCUGUUCCGGA__AACGAUUGUUUAAAAUUCCUUAGUUUGGCAGCUUAGAGCGACAAA .....(((((((((((((((((((.(((((......)))))....))))))....))).)))))))....)))....((((...((.....))..)))) (-18.91 = -19.72 + 0.81)

| Location | 16,161,332 – 16,161,429 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 90.24 |
| Shannon entropy | 0.19284 |
| G+C content | 0.44555 |
| Mean single sequence MFE | -23.93 |
| Consensus MFE | -22.11 |
| Energy contribution | -22.20 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.772594 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
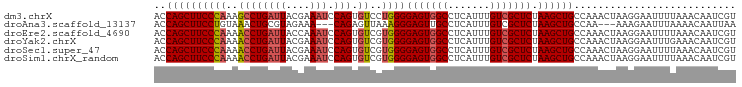
>dm3.chrX 16161332 97 - 22422827 ACCAGCUUCCCAAAGCCUGAUUACGAAAUCCAGUGUCCUGGGGAGUGGCCUCAUUUGUCGCUCUAAGCUGCCAAACUAAGGAAUUUUAAACAAUCGU ..((((((((((..((((((((....))).))).))..))))(((((((.......))))))).))))))((.......))................ ( -24.80, z-score = -1.19, R) >droAna3.scaffold_13137 108141 91 + 1373759 ACCAGCUUCCUGUAAACUGCGUAGAAA---CAGAGUUAAAGGGAGUUGCCUCAUUUGUCGCUCUAAGCUGCCAA---AAAGAAUUUAAAACAAUUAA ..(((((((((...((((..((....)---)..))))..))))))))).....((((.(((.....)).).)))---)................... ( -16.40, z-score = 0.11, R) >droEre2.scaffold_4690 7822858 97 + 18748788 ACCAGCUUCCCAAAACCUGAUUACCAAAUCCAGUGUCGUGGGGAGUGGCCUCAUUUGUCGCUCUAAGCUGCCAAACUAAGGAAUUUUAAACAAUCGU ..((((((((((..((((((((....))).))).))..))))(((((((.......))))))).))))))((.......))................ ( -25.60, z-score = -1.90, R) >droYak2.chrX 10304950 97 - 21770863 ACCAGCUUCCCAAAACCUGAUUACGAAAUCCAGUGUCGUGGGGAGUGGCCUCAUUUGUCGCUCUAAGCUGCCAAACUAAGGAAUUUGAAACAAUCGU ..((((((((((..((((((((....))).))).))..))))(((((((.......))))))).))))))((.......))................ ( -25.60, z-score = -1.38, R) >droSec1.super_47 209556 97 - 216421 ACCAGCUUCCCAAAACCUGAUUACGAAAUCCAGUGUCGUGGGGAGUGGCCUCAUUUGUCGCUCUAAGCUGCCAAACUAAGGAAUUUUAAACAAUCGU ..((((((((((..((((((((....))).))).))..))))(((((((.......))))))).))))))((.......))................ ( -25.60, z-score = -1.67, R) >droSim1.chrX_random 4204592 97 - 5698898 ACCAGCUUCCCAAAACCUGAUUACGAAAUCCAGUGUCGUGGGGAGUGGCCUCAUUUGUCGCUCUAAGCUGCCAAACUAAGGAAUUUUAAACAAUCGU ..((((((((((..((((((((....))).))).))..))))(((((((.......))))))).))))))((.......))................ ( -25.60, z-score = -1.67, R) >consensus ACCAGCUUCCCAAAACCUGAUUACGAAAUCCAGUGUCGUGGGGAGUGGCCUCAUUUGUCGCUCUAAGCUGCCAAACUAAGGAAUUUUAAACAAUCGU ..((((((((((..(((((...........))).))..))))(((((((.......))))))).))))))........................... (-22.11 = -22.20 + 0.09)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:49:10 2011