
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,036,946 – 1,037,105 |
| Length | 159 |
| Max. P | 0.523259 |

| Location | 1,036,946 – 1,037,105 |
|---|---|
| Length | 159 |
| Sequences | 5 |
| Columns | 168 |
| Reading direction | reverse |
| Mean pairwise identity | 65.25 |
| Shannon entropy | 0.61350 |
| G+C content | 0.42650 |
| Mean single sequence MFE | -40.74 |
| Consensus MFE | -15.22 |
| Energy contribution | -18.18 |
| Covariance contribution | 2.96 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.523259 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
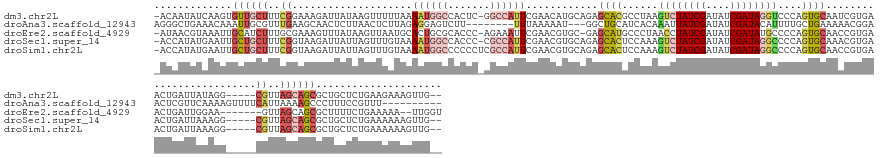
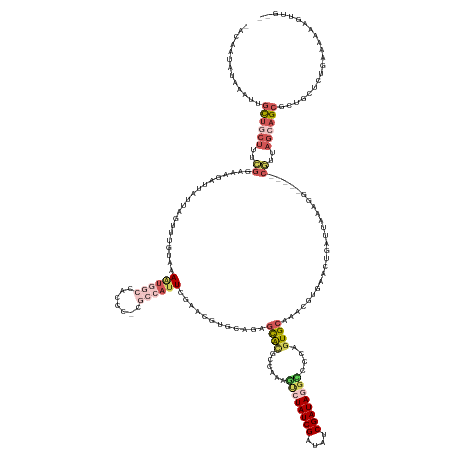
>dm3.chr2L 1036946 159 - 23011544 -ACAAUAUCAAGUGUUGCUUUCGGAAAGAUUAUAAGUUUUUAAAAUGGCCACUC-GGCCAUUCGAACAUGCAGAGCACGCCUAAGUCUAUCGAUAUCGAUAGGUCCCAGUGCAAUCGUGAACUGAUUAUAGG-----CGUUAGCAGCGCUGCUCUGAAGAAAGUUG-- -.((((((...))))))(((((.............((((....(((((((....-))))))).))))...(((((((((((((..(((((((....)))))))...((((.((....)).))))....))))-----))).(((...)))))))))..)))))...-- ( -49.90, z-score = -2.76, R) >droAna3.scaffold_12943 3549865 147 - 5039921 AGGGCUGAAACAAAUUGCGUUUGAAGCAACUCUUAACUCUUAGAGGAGUCUU--------UUUAAAAAU---GGCUGCAUCACAAAUUAUCGAUAUCGAUACAUUUUGCUGAAAAACGGAACUCGUUCAAAAGUUUUCAUUAAAAGCCCUUUCCGUUU---------- (((((((((((......(((((..(((((((((........)))).((((..--------.........---))))...........(((((....)))))....)))))...)))))(((....)))....))))).......))))))........---------- ( -31.41, z-score = -1.00, R) >droEre2.scaffold_4929 1078975 156 - 26641161 -AUAACGUAAAUUGCAUCUUUGCGAAAGUUUAUAAGUUAAUGCACUGCGCACCC-AGAAAUUCGAACGUGC-GAGCAUGCCCUAACCUAUCGAUAUCGAUAUGCCCCAGUGCAACCGUGAACUGAUUGGAA-------GUUAGCAGCGCUUUUCUGAAAAA--UUGGU -..........(((((....))))).(((((((..((...((((((((((((..-.(.....)....))))-).(((..........(((((....))))))))..))))))))).)))))))..(..(((-------(..(((...)))))))..)....--..... ( -33.30, z-score = 0.44, R) >droSec1.super_14 990416 159 - 2068291 -ACCAUAUGAAUUGCUGCUUUCGGUAAGAUUAUUAGUUUGUAAAAUGGCCACCC-CGCCAUUCGAACGUGCAGAGCACUCCAAAGUCUAUCGAUAUCGAUAGGCCCCAGUGCAAACGUGAACUGAUUAAAGG-----CGUUAGCAGCGCUGCUCUGAAAAAAGUUG-- -............((((((..((.(......((((((((....((((((.....-.))))))...((((.....(((((.....((((((((....))))))))...)))))..))))))))))))....).-----))..))))))...................-- ( -45.20, z-score = -2.03, R) >droSim1.chr2L 1007468 160 - 22036055 -ACCAUAUGAAUUGCUGCUUUCGGUAAGAUUAUUAGUUUGUAAAAUGGCCCCCCUCGCCAUUCGAACGUGCAGAGCACUCCAAAGUCUAUCGAUAUCGAUAGGCCCCAGUGCAACCGUGAACUGAUUAAAGG-----CGUUAGCAGCGCUGCUCUGAAAAAAGUUG-- -............((((((..((.(......(((((((..(..((((((.......))))))............(((((.....((((((((....))))))))...)))))....)..)))))))....).-----))..))))))...................-- ( -43.90, z-score = -1.67, R) >consensus _ACAAUAUAAAUUGCUGCUUUCGGAAAGAUUAUUAGUUUGUAAAAUGGCCACCC_CGCCAUUCGAACGUGCAGAGCACGCCAAAGUCUAUCGAUAUCGAUAGGCCCCAGUGCAAACGUGAACUGAUUAAAGG_____CGUUAGCAGCGCUGCUCUGAAAAAAGUUG__ .............((((((..((....................((((((.......))))))............((((......((((((((....))))))))....)))).........................))..))))))..................... (-15.22 = -18.18 + 2.96)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:07:34 2011