| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,138,087 – 16,138,180 |
| Length | 93 |
| Max. P | 0.789510 |

| Location | 16,138,087 – 16,138,180 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 75.52 |
| Shannon entropy | 0.41246 |
| G+C content | 0.48135 |
| Mean single sequence MFE | -22.13 |
| Consensus MFE | -13.06 |
| Energy contribution | -14.37 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.789510 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
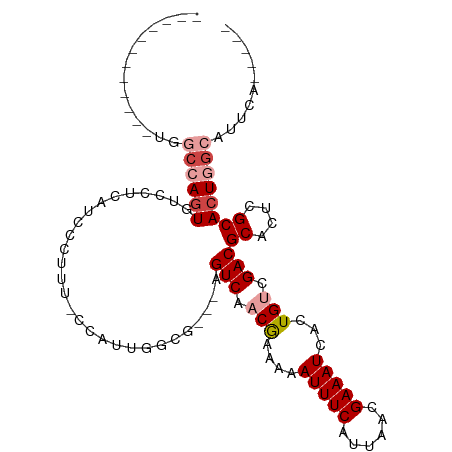
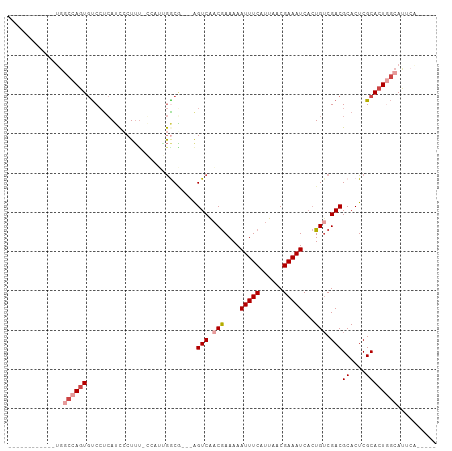
>dm3.chrX 16138087 93 + 22422827 ------------UGGCCAGUGUCCUCAUCCCUUU-GCACUGGCG---AGUCAACGAAAAAUUUCAUUAACGAAAUCACUGUCGACGCACUCGCACUGGCAUGGCAUUCA ------------..((((((((............-((....))(---((((..(((...(((((......))))).....)))..).)))))))))))).......... ( -24.90, z-score = -1.39, R) >droEre2.scaffold_4690 7797986 88 - 18748788 ------------UGUCCAGUGUCCUCAUCCCUUC-GCAAUGACG---GGUCGUCGAAAAAUUUCAUUAACGAAAUCACUGUCGACGCACUCGCACUCGCACUCG----- ------------.....(((((..((((......-...))))((---(((((((((...(((((......))))).....)))))).))))).....)))))..----- ( -20.90, z-score = -1.46, R) >droYak2.chrX 10277869 89 + 21770863 ------------UGGCCAGUGUCCUCAUCCCUUUUGCCCAGACG---AGUCAACGAAAAAUUUCAUUAACGAAAUCACUGUCGACGCACUCGCACUGGCAUUCA----- ------------..((((((((...((.......)).......(---((((..(((...(((((......))))).....)))..).)))))))))))).....----- ( -21.80, z-score = -1.91, R) >droSec1.super_47 186570 89 + 216421 ------------UGGCCAGUGUCCUCAUCCCUUUGCACUGGGCA---AGUCAACGAAAAAUUUCAUUAACGAAAUCACUGUCGACGCACUCGCACUGGCAUUCA----- ------------..((((((((......(((........)))..---.(((.(((....(((((......)))))...))).)))......)))))))).....----- ( -24.50, z-score = -2.31, R) >droSim1.chrX_random 4181892 89 + 5698898 ------------UGGCCAGUGUCCUCAUCCCUUUGCACUGGGCA---AGUCAACGAAAAAUUUCAUUAACGAAAUCACUGUCGACGCACUCGCACUGGCAUUCA----- ------------..((((((((......(((........)))..---.(((.(((....(((((......)))))...))).)))......)))))))).....----- ( -24.50, z-score = -2.31, R) >droPer1.super_22 439431 104 + 1688296 UGGCCAUUGCAACCCCUACUACCACGAGUACUACUCCAUUAGCUUGCAGUCGACAAAAAAUUUCAUUAACGAAAUCUGUGACGACGCACUCGCAUUCAAAUUCA----- .......(((...............(((.....))).....(..(((.((((.((....(((((......)))))...)).)))))))..))))..........----- ( -16.20, z-score = -0.77, R) >consensus ____________UGGCCAGUGUCCUCAUCCCUUU_CCAUUGGCG___AGUCAACGAAAAAUUUCAUUAACGAAAUCACUGUCGACGCACUCGCACUGGCAUUCA_____ ..............((((((((..................(((.....)))..(((...(((((......))))).....)))........)))))))).......... (-13.06 = -14.37 + 1.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:49:01 2011