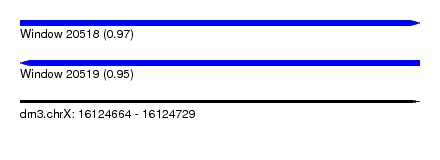
| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,124,664 – 16,124,729 |
| Length | 65 |
| Max. P | 0.973565 |

| Location | 16,124,664 – 16,124,729 |
|---|---|
| Length | 65 |
| Sequences | 3 |
| Columns | 65 |
| Reading direction | forward |
| Mean pairwise identity | 57.95 |
| Shannon entropy | 0.60477 |
| G+C content | 0.44242 |
| Mean single sequence MFE | -15.57 |
| Consensus MFE | -12.03 |
| Energy contribution | -11.27 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.64 |
| Mean z-score | -0.97 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.973565 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
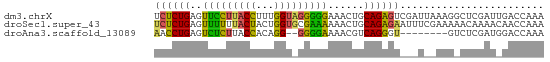
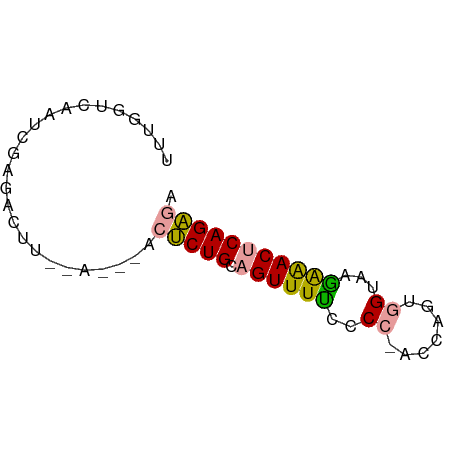
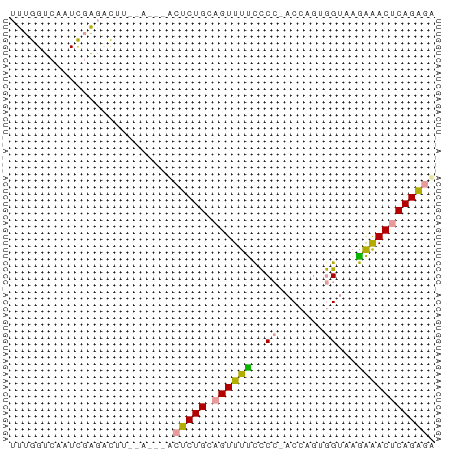
>dm3.chrX 16124664 65 + 22422827 UCUCUGAGUUCCUUACCUUUGGUAGGGGGAAACUGCAGAGUCGAUUAAAGGCUCGAUUGACCAAA ..(((((((((((((((...))))))))...))).))))(((((((........))))))).... ( -19.70, z-score = -1.28, R) >droSec1.super_43 270810 65 - 327307 UCUCUGAGUUUUUUACUACUGGUGCGAAAAAACUGCAGAGAAUUUCGAAAAACAAAACAACCAAA ((((((((((((((..((....))..)))))))).))))))........................ ( -13.30, z-score = -2.39, R) >droAna3.scaffold_13089 554769 55 + 978470 AACCUGAGUCUCUUACCACAGG--GGGGAAAACGUCAGGGU--------GUCUCGAUGGACCAAA ..(((((((.(((..((...))--..)))..)).)))))(.--------((((....)))))... ( -13.70, z-score = 0.78, R) >consensus UCUCUGAGUUUCUUACCACUGGU_GGGGAAAACUGCAGAGU___U__AAGACUCGAUCGACCAAA .(((((..(((((((((...)))))))))......)))))......................... (-12.03 = -11.27 + -0.77)
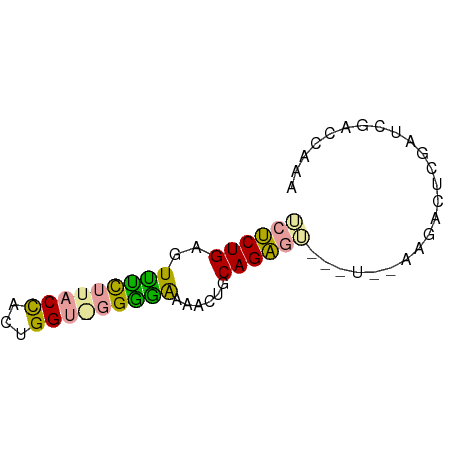
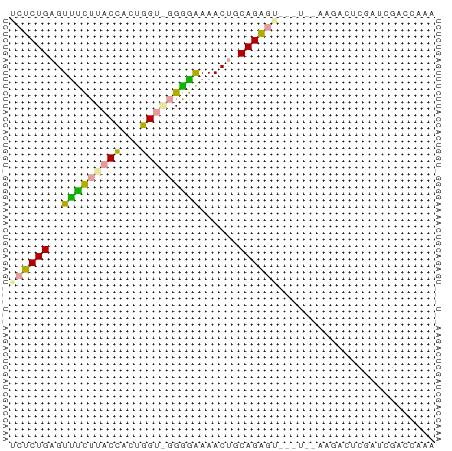
| Location | 16,124,664 – 16,124,729 |
|---|---|
| Length | 65 |
| Sequences | 3 |
| Columns | 65 |
| Reading direction | reverse |
| Mean pairwise identity | 57.95 |
| Shannon entropy | 0.60477 |
| G+C content | 0.44242 |
| Mean single sequence MFE | -15.07 |
| Consensus MFE | -8.33 |
| Energy contribution | -8.00 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.946643 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 16124664 65 - 22422827 UUUGGUCAAUCGAGCCUUUAAUCGACUCUGCAGUUUCCCCCUACCAAAGGUAAGGAACUCAGAGA .........((((........))))(((((.((((((..(((.....)))...))))))))))). ( -17.90, z-score = -1.92, R) >droSec1.super_43 270810 65 + 327307 UUUGGUUGUUUUGUUUUUCGAAAUUCUCUGCAGUUUUUUCGCACCAGUAGUAAAAAACUCAGAGA .......((((((.....))))))((((((.((((((((.((.......)))))))))))))))) ( -15.10, z-score = -2.19, R) >droAna3.scaffold_13089 554769 55 - 978470 UUUGGUCCAUCGAGAC--------ACCCUGACGUUUUCCCC--CCUGUGGUAAGAGACUCAGGUU ....(((......)))--------..(((((.(((((...(--(....))...)))))))))).. ( -12.20, z-score = 0.22, R) >consensus UUUGGUCAAUCGAGACUU__A___ACUCUGCAGUUUUCCCC_ACCAGUGGUAAGAAACUCAGAGA .........................(((((.((((((..((.......))...))))))))))). ( -8.33 = -8.00 + -0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:48:59 2011