
| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,098,958 – 16,099,070 |
| Length | 112 |
| Max. P | 0.773564 |

| Location | 16,098,958 – 16,099,070 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 79.13 |
| Shannon entropy | 0.29214 |
| G+C content | 0.48493 |
| Mean single sequence MFE | -35.25 |
| Consensus MFE | -17.95 |
| Energy contribution | -16.41 |
| Covariance contribution | -1.54 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.716013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 16098958 112 + 22422827 GCAUCCG--AACAGACGUUCGAUAUACGAGUAUGUGGGACUUUGAAGGAAUCGCGAUGGGUCGCGUU-UCCCCUACUCCUC-CCAUCUCCUUCACUCGUGUCUAAAUUGAUAUUUU ..(((((--(((....)))))..((((((((..((((((.......((((.(((((....))))).)-)))........))-)))).......)))))))).......)))..... ( -32.16, z-score = -1.76, R) >droYak2.chrX 10245916 102 + 21770863 GCAUCCG--AACAGACGUUCGAUAUGCGAGUGUGUGGUAGUUGGAAGGAAUCGUGGAGGGCUGCCUCCUCCUUCAC------------CAUCCACUCGUGUCUAAAUUGAUAUUUU ((((.((--(((....)))))..))))(((((..((((.....((((((.....(((((....)))))))))))))------------))..)))))(((((......)))))... ( -39.70, z-score = -3.85, R) >droSec1.super_47 155794 116 + 216421 GCAUCCAGCAACAGACGUUCGAUAUACGAGUAUGUGGGACUUUGAAGGAAUCGCGGUGGGCCGCUUCCUCCUCCAUCGCUCUCCUUCUCCUUCACUCGUGUCUAAAUUGAUAUUUU ((.....)).........(((((((((((((....((((....((((((...((((((((...........))))))))..))))))))))..))))))))....)))))...... ( -33.90, z-score = -1.89, R) >consensus GCAUCCG__AACAGACGUUCGAUAUACGAGUAUGUGGGACUUUGAAGGAAUCGCGGUGGGCCGCCUCCUCCUCCAC__CUC_CC_UCUCCUUCACUCGUGUCUAAAUUGAUAUUUU ..................(((((((((((((..(((((........(((...((((....))))....)))))))).................))))))))....)))))...... (-17.95 = -16.41 + -1.54)

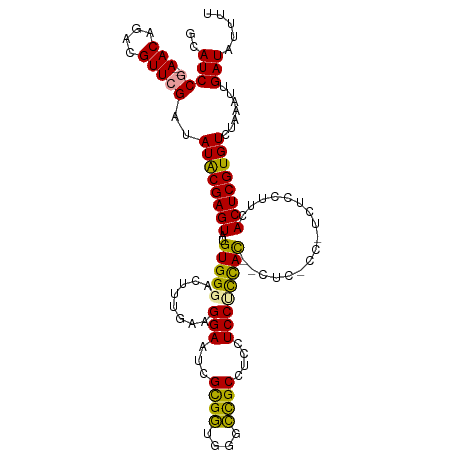
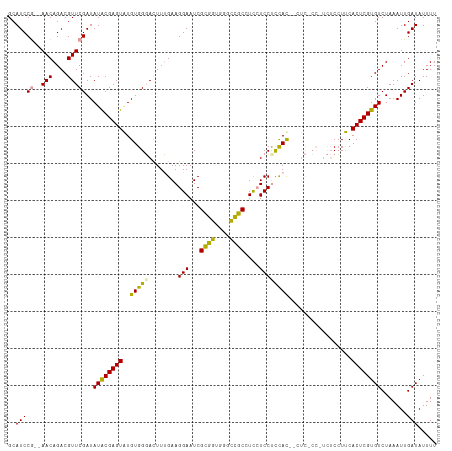
| Location | 16,098,958 – 16,099,070 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 79.13 |
| Shannon entropy | 0.29214 |
| G+C content | 0.48493 |
| Mean single sequence MFE | -34.87 |
| Consensus MFE | -18.84 |
| Energy contribution | -19.73 |
| Covariance contribution | 0.90 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.773564 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 16098958 112 - 22422827 AAAAUAUCAAUUUAGACACGAGUGAAGGAGAUGG-GAGGAGUAGGGGA-AACGCGACCCAUCGCGAUUCCUUCAAAGUCCCACAUACUCGUAUAUCGAACGUCUGUU--CGGAUGC ....((((.........(((((((..((.(((.(-(((((((...(..-..)((((....)))).))))))))...)))))...)))))))....(((((....)))--)))))). ( -36.70, z-score = -2.66, R) >droYak2.chrX 10245916 102 - 21770863 AAAAUAUCAAUUUAGACACGAGUGGAUG------------GUGAAGGAGGAGGCAGCCCUCCACGAUUCCUUCCAACUACCACACACUCGCAUAUCGAACGUCUGUU--CGGAUGC ...................(((((..((------------(((((((((((((....))))).....)))))).....))))..)))))((((.((((((....)))--))))))) ( -36.10, z-score = -3.91, R) >droSec1.super_47 155794 116 - 216421 AAAAUAUCAAUUUAGACACGAGUGAAGGAGAAGGAGAGCGAUGGAGGAGGAAGCGGCCCACCGCGAUUCCUUCAAAGUCCCACAUACUCGUAUAUCGAACGUCUGUUGCUGGAUGC ....((((....((((((((((((..((((((((((.(((.(((.(..(....)..)))).)))..)))))))....)))....))))))).........)))))......)))). ( -31.80, z-score = -0.82, R) >consensus AAAAUAUCAAUUUAGACACGAGUGAAGGAGA_GG_GAG__GUGGAGGAGGAAGCGGCCCACCGCGAUUCCUUCAAAGUCCCACAUACUCGUAUAUCGAACGUCUGUU__CGGAUGC ....((((....((((((((((((..................(((((((...((((....))))..)))))))...(.....).))))))).........)))))......)))). (-18.84 = -19.73 + 0.90)

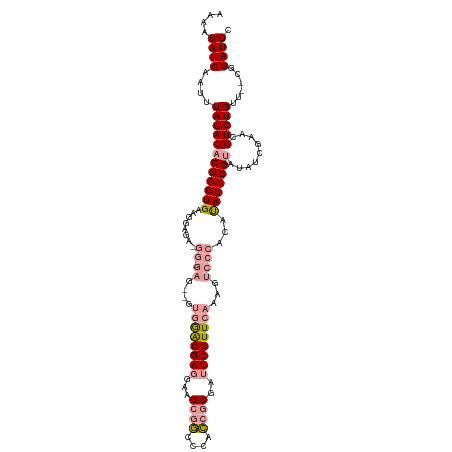
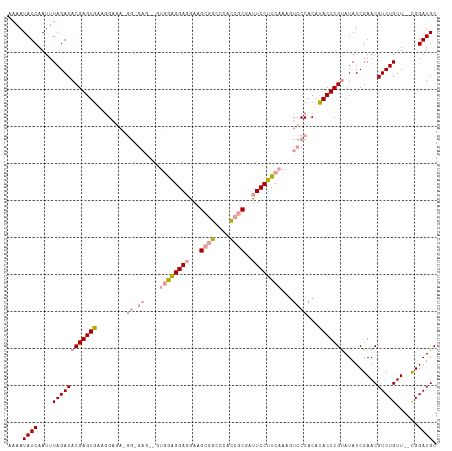
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:48:57 2011