| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,496,120 – 11,496,210 |
| Length | 90 |
| Max. P | 0.655753 |

| Location | 11,496,120 – 11,496,210 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 75.25 |
| Shannon entropy | 0.48092 |
| G+C content | 0.46099 |
| Mean single sequence MFE | -25.36 |
| Consensus MFE | -12.39 |
| Energy contribution | -12.93 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.655753 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
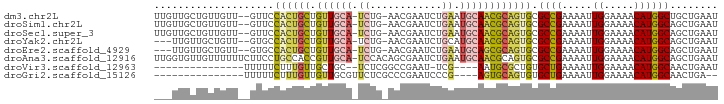
>dm3.chr2L 11496120 90 - 23011544 UUGUUGCUGUUGUU--GUUCCACUGCUGUUGCA-UCUG-AACGAAUCUGAAUGCAACGCAGUGCGCCGAAAAUUGGAAAACAUGGCUGCUGAAU ..((.(((((.(((--....((((((.((((((-((.(-(.....)).).)))))))))))))..((((...))))..)))))))).))..... ( -25.90, z-score = -1.76, R) >droSim1.chr2L 11310366 90 - 22036055 UUGUUGCUGUUGUU--GUUCCACUGCUGUUGCA-UCUG-AACGAAUCUGAAUGCAACGCAGUGCGCCGAAAAUUGGAAAACAUGGCAGCUGAAU .....(((((..(.--(((.((((((.((((((-((.(-(.....)).).)))))))))))))..((((...))))..))))..)))))..... ( -29.90, z-score = -2.64, R) >droSec1.super_3 6891183 90 - 7220098 UUGUUGCUGUUGUU--GUUCCACUGCUGUUGCA-UCUG-AACGAAUCUGAAUGCAACGCAGUGCGCCGAAAAUUGGAAAACAUGGCAGCUGAAU .....(((((..(.--(((.((((((.((((((-((.(-(.....)).).)))))))))))))..((((...))))..))))..)))))..... ( -29.90, z-score = -2.64, R) >droYak2.chr2L 7902649 87 - 22324452 ---UUGUUGCUGUU--GUGCCACUGCUGUUGCA-UCUG-AACGAAUCUGCAUGCAACGCAGUGCGCCGAAAAUUGGAAAACAUGGCAGCUGAAU ---.....(((((.--.((.((((((.((((((-((.(-(.....)).).)))))))))))))..((((...))))....))..)))))..... ( -31.60, z-score = -2.57, R) >droEre2.scaffold_4929 12704849 87 + 26641161 ---UUGUUGCUGUU--GUGCCACUGCUGUUGCA-UCUG-AACGAAUCUGAAUGCAGCGCAGUGCGCCGAAAAUUGGAAAACAUGGCAGCUGAAU ---.....(((((.--.((.((((((.((((((-((.(-(.....)).).)))))))))))))..((((...))))....))..)))))..... ( -31.60, z-score = -2.66, R) >droAna3.scaffold_12916 12195048 93 + 16180835 UUGGUGUUGUUUUUUCUUCCUGCCACCGUUGCA-UCCACAGCGAAUCUGAAUGCAACGCAGUGCGCCGAAAAUUGGAAAACAUGGCAGCUGAAU .............(((...((((((.(((((((-(...(((.....))).))))))))..((...((((...))))...)).))))))..))). ( -24.70, z-score = -0.23, R) >droVir3.scaffold_12963 18156392 72 + 20206255 ---------------UUUUUCUUUGUUGCUGC--UCUCGGCCGAAU-UCG----AAUGCGCUGUGCUGAAAAUUGGAAAACAUGGCAACUGAAU ---------------...(((...((((((((--..((((......-)))----)..))..(((.(..(...)..)...))).)))))).))). ( -14.60, z-score = -0.27, R) >droGri2.scaffold_15126 7337777 73 + 8399593 ---------------UUUUUCUUUGUUGUUGCGUUCUCGCCCGAAUCCCG----AGUGCAGUGUGCUGAAAAUUGGAAAACAUGGCAACUGA-- ---------------.........(((((((((....))).......(((----(...(((....)))....)))).......))))))...-- ( -14.70, z-score = 0.21, R) >consensus ___UUGCUGUUGUU__GUUCCACUGCUGUUGCA_UCUG_AACGAAUCUGAAUGCAACGCAGUGCGCCGAAAAUUGGAAAACAUGGCAGCUGAAU ....................((((((.((((((.((............)).)))))))))))).((((.....((.....))))))........ (-12.39 = -12.93 + 0.53)

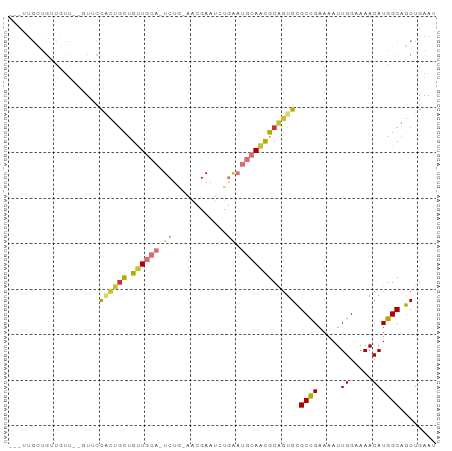
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:32:54 2011