
| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,077,219 – 16,077,427 |
| Length | 208 |
| Max. P | 0.871287 |

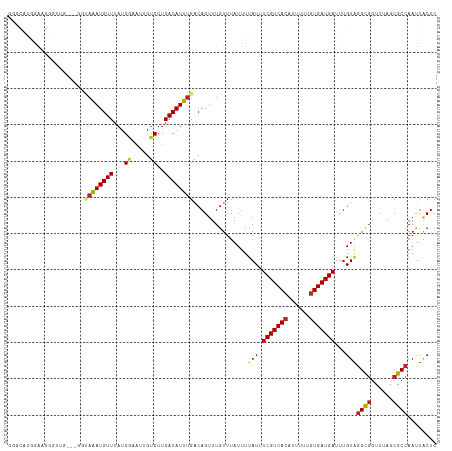
| Location | 16,077,219 – 16,077,337 |
|---|---|
| Length | 118 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 82.82 |
| Shannon entropy | 0.39048 |
| G+C content | 0.36148 |
| Mean single sequence MFE | -26.83 |
| Consensus MFE | -18.61 |
| Energy contribution | -18.80 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.871287 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 16077219 118 + 22422827 GGGCAUGGAAGGGUUGUGUUGUAAAUGUUUAUGGUAGUGUCUUGACAUUUGACAGUUUGUUUAUUUUAUUUCGUCACAUUUUUGUGAUGAUUUGUAGGCGGUUUAGCGCCAAUUACUC .(((..............(((((((((((...((......)).))))))).)))).((((((((......((((((((....))))))))...))))))))......)))........ ( -26.60, z-score = -0.92, R) >droYak2.chrX 10225319 113 + 21770863 GGGCAUGGAAGGGUUG-----UAAAUGUUUAUGGUAGUGUCUUGACAUUUGACAGUUUGUUUAUUUUAUUUCGUCACAUUUUUGUGAUGAUUUGUAGGCGGUUUAGCGCCAAUUACUC .((((((((.(..(((-----((((((((...((......)).))))))).))))..).))))).((((.((((((((....))))))))...))))((......)))))........ ( -27.50, z-score = -1.72, R) >droEre2.scaffold_4690 7744596 113 - 18748788 GGGCCUGGAGGGGUUG-----UAAAUGUUUAUGGUAGUGUCUUGACAUUUGACAGUUUGUUUAUUUUAUUUCGUCACAUUUUUGUGAUGAUUUGUAGGCGGUUUAGCGCCAAUUACUC .((((((((.(..(((-----((((((((...((......)).))))))).))))..)((((((......((((((((....))))))))...))))))..))))).)))........ ( -30.40, z-score = -2.30, R) >droSec1.super_47 134704 118 + 216421 GGGCAUGGAAGGGUUGUGUUGUAAAUGUUUAUGGUAGUGUCUUGACAUUUGACAGUUUGUUUAUUUUAUUUCGUCACAUUUUUGUGAUGAUUUGUAGGCGGUUUAGCGCCAAUUACUC .(((..............(((((((((((...((......)).))))))).)))).((((((((......((((((((....))))))))...))))))))......)))........ ( -26.60, z-score = -0.92, R) >droSim1.chrX 12417913 118 + 17042790 GGGCAUGGAAGGGUUGUGUUGUAAAUGUUUAUGGUAGUGUCUUGACAUUUGACAGUUUGUUUAUUUUAUUUCGUCACAUUUUUGUGAUGAUUUGUAGGCGGUUUAGCGCCAAUUACUC .(((..............(((((((((((...((......)).))))))).)))).((((((((......((((((((....))))))))...))))))))......)))........ ( -26.60, z-score = -0.92, R) >droMoj3.scaffold_6308 3098825 110 + 3356042 ----GGGAAUCGGAGG---GACAGAUGUUUAUGAA-UCGUCUUGACAUUUGACAGUUUGUUUAUUUUAUUUCGUCACAUUUUCGUGAUGAUUUGUAGGCGGUUUAGCGCCAAUUACUC ----..(((.((((..---(.((((((((...((.-...))..)))))))).)..)))))))....((..(((((((......)))))))..))..((((......))))........ ( -25.20, z-score = -0.77, R) >droGri2.scaffold_15203 4588634 114 + 11997470 AAGUAUAAAAGACGGG---GACAAAUGUUUAUGAA-UAGUCUUGACAUUUGACAGCUUGUUUAUUUUAUUUCGUCACAUUUUCGUGAUGAUUUGUAGGCGGUUUAGCGCCAAUUACUC ...(((((((((((((---..((((((((...((.-...))..))))))))....)))))))........(((((((......)))))))))))))((((......))))........ ( -29.00, z-score = -2.44, R) >droVir3.scaffold_12472 382458 96 - 763072 ----------------------AGAUGUUUAUGAAAUAGUCUUGACAUUUGACAGUUUGUUUAUUUUAUUUCGUCACAUUUUCGUGAUGAUUUGUGGGCGGUUUAGCGCCAAUUACUC ----------------------........(((((((((....(((........)))...))))))))).(((((((......)))))))......((((......))))........ ( -19.80, z-score = -0.96, R) >droWil1.scaffold_181096 6835364 100 - 12416693 ------------------UGGGAAAUGUUUAUGGAAUUGUCUUGACAUUUGACAGUUUGUUUAUUUUAUUUCGUCACAUUUUUGUGAUGAUUUGUAGGCGGUUUAGCGCCAAUUACUC ------------------((..(((((((...(((....))).)))))))..))............((..((((((((....))))))))..))..((((......))))........ ( -25.10, z-score = -2.48, R) >droPer1.super_22 1187691 118 - 1688296 AGACAGAAGAGUGGUAAAUGGUAAAUGUUUAUGGAAUAGUCUUGACAUUUGACAGUUUGUUUAUUUUAUUUCGUCACAUUUUUGUGAUGAUUUGUAGGCGGUUUAGCGCCAAUUACUC ........(((((((((((..((((((((...(((....))).))))))))...))))........((..((((((((....))))))))..))..((((......)))).))))))) ( -30.60, z-score = -2.66, R) >dp4.chrXL_group1e 9705177 118 + 12523060 AGACAGAAGAGUGGUAAAUGGUAAAUGUUUAUGGAAUAGUCUUGACAUUUGACAGUUUGUUUAUUUUAUUUCGUCACAUUUUUGUGAUGAUUUGUAGGCGGUUUAGCGCCAAUUACUC ........(((((((((((..((((((((...(((....))).))))))))...))))........((..((((((((....))))))))..))..((((......)))).))))))) ( -30.60, z-score = -2.66, R) >droAna3.scaffold_13260 1939045 110 + 2125536 GCGUAUAGGGAAUCUG------UAAUGUUUAUGGAAAAGACUUGACAUUUGGCAGUUUGUUUAUUUUAUUUCGUCACAUUUUGUUGAUGAU--GUUAGUCUUUUAGCGCCAACGGCUA ((.(((((.....)))------))..........((((((((.((((((..((((..(((...............)))..))))..)..))--))))))))))).))(((...))).. ( -23.96, z-score = -0.71, R) >consensus GGGCAUGGAAGGGUUG___GGUAAAUGUUUAUGGAAUUGUCUUGACAUUUGACAGUUUGUUUAUUUUAUUUCGUCACAUUUUUGUGAUGAUUUGUAGGCGGUUUAGCGCCAAUUACUC .....................((((((((...((.....))..))))))))...................(((((((......)))))))......((((......))))........ (-18.61 = -18.80 + 0.19)


| Location | 16,077,219 – 16,077,337 |
|---|---|
| Length | 118 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 82.82 |
| Shannon entropy | 0.39048 |
| G+C content | 0.36148 |
| Mean single sequence MFE | -14.21 |
| Consensus MFE | -10.74 |
| Energy contribution | -10.93 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.659856 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
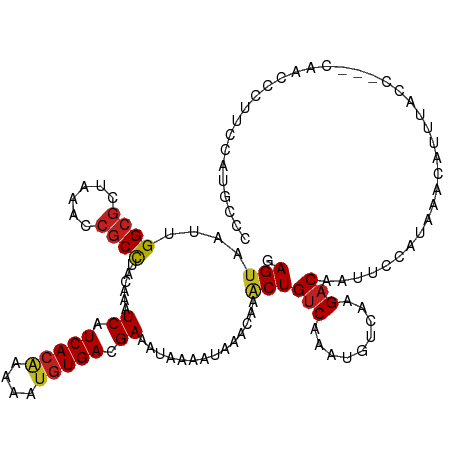
>dm3.chrX 16077219 118 - 22422827 GAGUAAUUGGCGCUAAACCGCCUACAAAUCAUCACAAAAAUGUGACGAAAUAAAAUAAACAAACUGUCAAAUGUCAAGACACUACCAUAAACAUUUACAACACAACCCUUCCAUGCCC ..((((..((((......))))......((.(((((....))))).))................((((.........)))).............)))).................... ( -14.50, z-score = -1.37, R) >droYak2.chrX 10225319 113 - 21770863 GAGUAAUUGGCGCUAAACCGCCUACAAAUCAUCACAAAAAUGUGACGAAAUAAAAUAAACAAACUGUCAAAUGUCAAGACACUACCAUAAACAUUUA-----CAACCCUUCCAUGCCC ..((((..((((......))))......((.(((((....))))).))................((((.........)))).............)))-----)............... ( -14.50, z-score = -1.46, R) >droEre2.scaffold_4690 7744596 113 + 18748788 GAGUAAUUGGCGCUAAACCGCCUACAAAUCAUCACAAAAAUGUGACGAAAUAAAAUAAACAAACUGUCAAAUGUCAAGACACUACCAUAAACAUUUA-----CAACCCCUCCAGGCCC (((.....((((......))))......((.(((((....))))).))................(((.((((((................)))))))-----))....)))....... ( -14.79, z-score = -1.10, R) >droSec1.super_47 134704 118 - 216421 GAGUAAUUGGCGCUAAACCGCCUACAAAUCAUCACAAAAAUGUGACGAAAUAAAAUAAACAAACUGUCAAAUGUCAAGACACUACCAUAAACAUUUACAACACAACCCUUCCAUGCCC ..((((..((((......))))......((.(((((....))))).))................((((.........)))).............)))).................... ( -14.50, z-score = -1.37, R) >droSim1.chrX 12417913 118 - 17042790 GAGUAAUUGGCGCUAAACCGCCUACAAAUCAUCACAAAAAUGUGACGAAAUAAAAUAAACAAACUGUCAAAUGUCAAGACACUACCAUAAACAUUUACAACACAACCCUUCCAUGCCC ..((((..((((......))))......((.(((((....))))).))................((((.........)))).............)))).................... ( -14.50, z-score = -1.37, R) >droMoj3.scaffold_6308 3098825 110 - 3356042 GAGUAAUUGGCGCUAAACCGCCUACAAAUCAUCACGAAAAUGUGACGAAAUAAAAUAAACAAACUGUCAAAUGUCAAGACGA-UUCAUAAACAUCUGUC---CCUCCGAUUCCC---- ..(((...((((......))))))).((((.....((.....(((((.................)))))....))..(((((-(........))).)))---.....))))...---- ( -13.83, z-score = -0.22, R) >droGri2.scaffold_15203 4588634 114 - 11997470 GAGUAAUUGGCGCUAAACCGCCUACAAAUCAUCACGAAAAUGUGACGAAAUAAAAUAAACAAGCUGUCAAAUGUCAAGACUA-UUCAUAAACAUUUGUC---CCCGUCUUUUAUACUU (((((...((((......))))......((.((((......)))).)).................(.(((((((...((...-.))....))))))).)---...........))))) ( -16.30, z-score = -0.77, R) >droVir3.scaffold_12472 382458 96 + 763072 GAGUAAUUGGCGCUAAACCGCCCACAAAUCAUCACGAAAAUGUGACGAAAUAAAAUAAACAAACUGUCAAAUGUCAAGACUAUUUCAUAAACAUCU---------------------- ((......((((......))))......)).((((......)))).((((((.............(((.........)))))))))..........---------------------- ( -11.51, z-score = -0.57, R) >droWil1.scaffold_181096 6835364 100 + 12416693 GAGUAAUUGGCGCUAAACCGCCUACAAAUCAUCACAAAAAUGUGACGAAAUAAAAUAAACAAACUGUCAAAUGUCAAGACAAUUCCAUAAACAUUUCCCA------------------ ((((....((((......))))......((.(((((....))))).))................((((.........))))))))...............------------------ ( -13.90, z-score = -1.80, R) >droPer1.super_22 1187691 118 + 1688296 GAGUAAUUGGCGCUAAACCGCCUACAAAUCAUCACAAAAAUGUGACGAAAUAAAAUAAACAAACUGUCAAAUGUCAAGACUAUUCCAUAAACAUUUACCAUUUACCACUCUUCUGUCU ((((....((((......))))......((.(((((....))))).))................((..((((((................))))))..))......))))........ ( -16.19, z-score = -1.79, R) >dp4.chrXL_group1e 9705177 118 - 12523060 GAGUAAUUGGCGCUAAACCGCCUACAAAUCAUCACAAAAAUGUGACGAAAUAAAAUAAACAAACUGUCAAAUGUCAAGACUAUUCCAUAAACAUUUACCAUUUACCACUCUUCUGUCU ((((....((((......))))......((.(((((....))))).))................((..((((((................))))))..))......))))........ ( -16.19, z-score = -1.79, R) >droAna3.scaffold_13260 1939045 110 - 2125536 UAGCCGUUGGCGCUAAAAGACUAAC--AUCAUCAACAAAAUGUGACGAAAUAAAAUAAACAAACUGCCAAAUGUCAAGUCUUUUCCAUAAACAUUA------CAGAUUCCCUAUACGC ..(((...)))...((((((((.((--((..........))))((((........................)))).))))))))............------................ ( -9.76, z-score = 0.94, R) >consensus GAGUAAUUGGCGCUAAACCGCCUACAAAUCAUCACAAAAAUGUGACGAAAUAAAAUAAACAAACUGUCAAAUGUCAAGACAAUUCCAUAAACAUUUACC___CAACCCUUCCAUGCCC .(((....((((......))))......((.(((((....))))).))..............)))(((.........)))...................................... (-10.74 = -10.93 + 0.19)

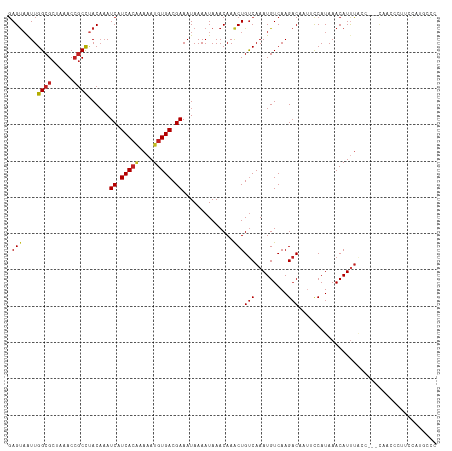
| Location | 16,077,257 – 16,077,372 |
|---|---|
| Length | 115 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.77 |
| Shannon entropy | 0.24840 |
| G+C content | 0.36541 |
| Mean single sequence MFE | -26.38 |
| Consensus MFE | -17.01 |
| Energy contribution | -17.41 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.598757 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 16077257 115 + 22422827 GUCUUGACAUUUGACAGUUUGUUUAUUUUAUUUCGUCACAUUUUUGUGAUGAUUUGUAGGCGGUUUAGCGCCAAUUACUCCACC-AAACUUUUACAAUGCAGACUCUGGUAAACGG---- ((((.(.(((((((.((((((.......((..((((((((....))))))))..))..((((......))))...........)-))))).))).))))))))).(((.....)))---- ( -29.70, z-score = -2.88, R) >droYak2.chrX 10225352 113 + 21770863 GUCUUGACAUUUGACAGUUUGUUUAUUUUAUUUCGUCACAUUUUUGUGAUGAUUUGUAGGCGGUUUAGCGCCAAUUACUCCACC-AAACUUUUACAAUGCAGAC--UGGUAAACGG---- ((((.(.(((((((.((((((.......((..((((((((....))))))))..))..((((......))))...........)-))))).))).)))))))))--..........---- ( -29.30, z-score = -2.78, R) >droEre2.scaffold_4690 7744629 113 - 18748788 GUCUUGACAUUUGACAGUUUGUUUAUUUUAUUUCGUCACAUUUUUGUGAUGAUUUGUAGGCGGUUUAGCGCCAAUUACUCCACC-AAACUUUUACGCUGCAGAC--UGGUAAACGG---- .......(.((((.((((((((......((..((((((((....))))))))..))..((((......))))............-.............))))))--)).)))).).---- ( -27.70, z-score = -1.96, R) >droSec1.super_47 134742 115 + 216421 GUCUUGACAUUUGACAGUUUGUUUAUUUUAUUUCGUCACAUUUUUGUGAUGAUUUGUAGGCGGUUUAGCGCCAAUUACUCCACC-AAACUUUUACAAUGCAGUCUCUGGUAAACGG---- ...(((.(((((((.((((((.......((..((((((((....))))))))..))..((((......))))...........)-))))).))).)))))))...(((.....)))---- ( -26.40, z-score = -1.79, R) >droSim1.chrX 12417951 115 + 17042790 GUCUUGACAUUUGACAGUUUGUUUAUUUUAUUUCGUCACAUUUUUGUGAUGAUUUGUAGGCGGUUUAGCGCCAAUUACUCCACC-AAACUUUUACAAUGCAGUCUCUGGUAAACGG---- ...(((.(((((((.((((((.......((..((((((((....))))))))..))..((((......))))...........)-))))).))).)))))))...(((.....)))---- ( -26.40, z-score = -1.79, R) >droMoj3.scaffold_6308 3098855 108 + 3356042 GUCUUGACAUUUGACAGUUUGUUUAUUUUAUUUCGUCACAUUUUCGUGAUGAUUUGUAGGCGGUUUAGCGCCAAUUACUCCAC---AACUUUUACAAUGCAGAC--UG-UAAAC------ .............(((((((((......((..(((((((......)))))))..))..((((......))))...........---............))))))--))-)....------ ( -26.60, z-score = -2.74, R) >droGri2.scaffold_15203 4588668 108 + 11997470 GUCUUGACAUUUGACAGCUUGUUUAUUUUAUUUCGUCACAUUUUCGUGAUGAUUUGUAGGCGGUUUAGCGCCAAUUACUCCAC---GAAUUUCAAAAUGCAGAC--UG-UAAAC------ ((((.(.(((((......((((......((..(((((((......)))))))..))..((((......)))).........))---))......))))))))))--..-.....------ ( -23.70, z-score = -1.27, R) >droVir3.scaffold_12472 382474 108 - 763072 GUCUUGACAUUUGACAGUUUGUUUAUUUUAUUUCGUCACAUUUUCGUGAUGAUUUGUGGGCGGUUUAGCGCCAAUUACUCCAC---AACUUUUACAAUGCAGAC--UG-UAAAC------ .............(((((((((..........(((((((......))))))).(((((((((......)))........))))---))..........))))))--))-)....------ ( -27.10, z-score = -2.53, R) >droWil1.scaffold_181096 6835384 108 - 12416693 GUCUUGACAUUUGACAGUUUGUUUAUUUUAUUUCGUCACAUUUUUGUGAUGAUUUGUAGGCGGUUUAGCGCCAAUUACUCCAC---AACUUUUACAGUCCAGAC--UG-UAAAC------ (((.........)))((((.((......((..((((((((....))))))))..))..((((......)))).........))---))))((((((((....))--))-)))).------ ( -27.50, z-score = -3.13, R) >droPer1.super_22 1187729 119 - 1688296 GUCUUGACAUUUGACAGUUUGUUUAUUUUAUUUCGUCACAUUUUUGUGAUGAUUUGUAGGCGGUUUAGCGCCAAUUACUCCACCGAAACUUUUACAAUGCAGCGACUG-UAAACUUUAAA ((((((.(((((((.(((((........((..((((((((....))))))))..))..((((......)))).............))))).))).))))))).)))..-........... ( -27.50, z-score = -2.10, R) >dp4.chrXL_group1e 9705215 119 + 12523060 GUCUUGACAUUUGACAGUUUGUUUAUUUUAUUUCGUCACAUUUUUGUGAUGAUUUGUAGGCGGUUUAGCGCCAAUUACUCCACCGAAACUUUUACAAUGCAGCGACUG-UAAACUUUAAA ((((((.(((((((.(((((........((..((((((((....))))))))..))..((((......)))).............))))).))).))))))).)))..-........... ( -27.50, z-score = -2.10, R) >droAna3.scaffold_13260 1939077 105 + 2125536 GACUUGACAUUUGGCAGUUUGUUUAUUUUAUUUCGUCACAUUUUGUUGAUGA--UGUUAGUCUUUUAGCGCCAACGGCUACAAU-AAGCAUUUUAAAUACAC----UGACAA-------- ..........(((.((((.((((((.......((((((........))))))--.(((((....)))))(((...)))......-........)))))).))----)).)))-------- ( -17.10, z-score = 0.17, R) >consensus GUCUUGACAUUUGACAGUUUGUUUAUUUUAUUUCGUCACAUUUUUGUGAUGAUUUGUAGGCGGUUUAGCGCCAAUUACUCCACC_AAACUUUUACAAUGCAGAC__UG_UAAAC______ ...(((.(((((((.((((.........((..(((((((......)))))))..))..((((......))))..............)))).))).))))))).................. (-17.01 = -17.41 + 0.40)



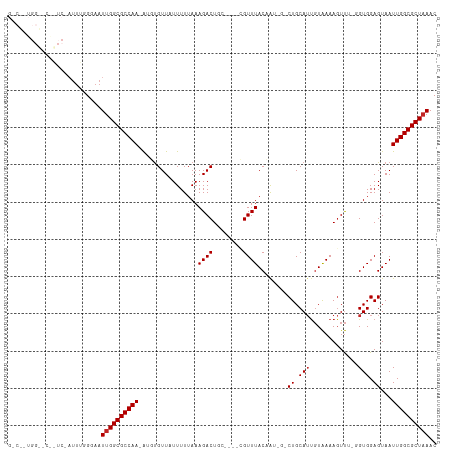
| Location | 16,077,319 – 16,077,427 |
|---|---|
| Length | 108 |
| Sequences | 10 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 76.91 |
| Shannon entropy | 0.44840 |
| G+C content | 0.41378 |
| Mean single sequence MFE | -28.19 |
| Consensus MFE | -16.49 |
| Energy contribution | -16.69 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.834508 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 16077319 108 - 22422827 GGCGUUGGCUCGGUC-AUUUGGUAAUUGGCGCCAA-AUGUGUUAUUUUUAAAGACUGC----CGUUUACCAGAGUCUGCAUUGUAAAAGUUU-GGUGGAGUAAUUGGCGCUAAAC (((((((((..((((-.(((((....((((((...-..))))))...)))))))))))----).(((((((((.(.(........).).)))-))))))......)))))).... ( -28.00, z-score = -0.36, R) >droYak2.chrX 10225414 106 - 21770863 GGCGGUGGCUAGGUC-AUUUGGUAAUUGGCGCCAA-AUGUGUUAUUUUUAAAGACUGC----CGUUUACCA--GUCUGCAUUGUAAAAGUUU-GGUGGAGUAAUUGGCGCUAAAC (((((((((...)))-))).(.((((((.((((((-((...((((......((((((.----.......))--)))).....))))..))))-))))...)))))).)))).... ( -27.40, z-score = -0.23, R) >droEre2.scaffold_4690 7744691 87 + 18748788 --------------------GGCAAUUGGCGCCAA-AUGCGUUAUUUUUAAAGACUGC----CGUUUACCA--GUCUGCAGCGUAAAAGUUU-GGUGGAGUAAUUGGCGCUAAAC --------------------.....((((((((((-.(((...........((((((.----.......))--)))).((.((........)-).))..))).)))))))))).. ( -27.20, z-score = -1.65, R) >droSec1.super_47 134804 108 - 216421 GUCGGUGGCUCGGUC-AUUUGGUAAUUGGCGCCAA-AUGUGUUAUUUUUAAAGACUGC----CGUUUACCAGAGACUGCAUUGUAAAAGUUU-GGUGGAGUAAUUGGCGCUAAAC ((((((.((((((((-.((((((((.(((((....-..((.((.......)).)))))----)).)))))))))))).(((((........)-)))))))).))))))....... ( -30.30, z-score = -1.25, R) >droSim1.chrX 12418013 109 - 17042790 GUCGGUGGCUCGGUCUUUUUGGUAAUUGGCGCCAA-AUGUGUUAUUUUUAAAGACUGC----CGUUUACCAGAGACUGCAUUGUAAAAGUUU-GGUGGAGUAAUUGGCGCUAAAC ((((((.((((((((((..((((((.(((((....-..((.((.......)).)))))----)).)))))))))))).(((((........)-)))))))).))))))....... ( -29.70, z-score = -1.03, R) >droMoj3.scaffold_6308 3098917 85 - 3356042 ----------------AUUAUGGAAUCGGCGCCAACA--UGUUCUUUUUAAAGACU------CGUUUACAGU---CUGCAUUGUAAAAGUU---GUGGAGUAAUUGGCGCUAAAC ----------------...........((((((((..--((((((.......((((------..((((((((---....))))))))))))---..)))))).)))))))).... ( -20.50, z-score = -1.33, R) >droGri2.scaffold_15203 4588730 85 - 11997470 ----------------AUUAAGGAAUUGGCGCCAACG--UGUUCUUUUUAAAGACU------AGUUUACAGU---CUGCAUUUUGAAAUUC---GUGGAGUAAUUGGCGCUAAAC ----------------.........((((((((.(((--..(((.......(((((------.(....))))---)).......)))...)---)).((....)))))))))).. ( -21.94, z-score = -1.94, R) >droVir3.scaffold_12472 382536 85 + 763072 ----------------AUUAAGGAAUUGGCGCCAACA--UGUUCUUUUUAAAGACU------AGUUUACAGU---CUGCAUUGUAAAAGUU---GUGGAGUAAUUGGCGCUAAAC ----------------.........((((((((((..--((((((.......((((------..((((((((---....))))))))))))---..)))))).)))))))))).. ( -22.50, z-score = -2.15, R) >droPer1.super_22 1187791 114 + 1688296 GGCUUUGGGCCAUUCCAUUGAGGAAUUGGCGCCAACAUUUGUUCUUUUUAAAGACUGUUUAAAGUUUACAGU-CGCUGCAUUGUAAAAGUUUCGGUGGAGUAAUUGGCGCUAAAC (((.....))).((((.....))))((((((((((.................((((((.........)))))-)(((.(((((.........))))).)))..)))))))))).. ( -37.20, z-score = -3.14, R) >dp4.chrXL_group1e 9705277 114 - 12523060 GGCUUUGGGCCAUUCCAUUGAGGAAUUGGCGCCAACAUUUGUUCUUUUUAAAGACUGUUUAAAGUUUACAGU-CGCUGCAUUGUAAAAGUUUCGGUGGAGUAAUUGGCGCUAAAC (((.....))).((((.....))))((((((((((.................((((((.........)))))-)(((.(((((.........))))).)))..)))))))))).. ( -37.20, z-score = -3.14, R) >consensus G_C__UGG__C__UC_AUUUGGGAAUUGGCGCCAA_AUGUGUUAUUUUUAAAGACUGC____CGUUUACAAU_G_CUGCAUUGUAAAAGUUU_GGUGGAGUAAUUGGCGCUAAAC .........................((((((((((................((((........))))........((.(((.............))).))...)))))))))).. (-16.49 = -16.69 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:48:53 2011