| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,068,315 – 16,068,409 |
| Length | 94 |
| Max. P | 0.947571 |

| Location | 16,068,315 – 16,068,409 |
|---|---|
| Length | 94 |
| Sequences | 11 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 70.07 |
| Shannon entropy | 0.55443 |
| G+C content | 0.45367 |
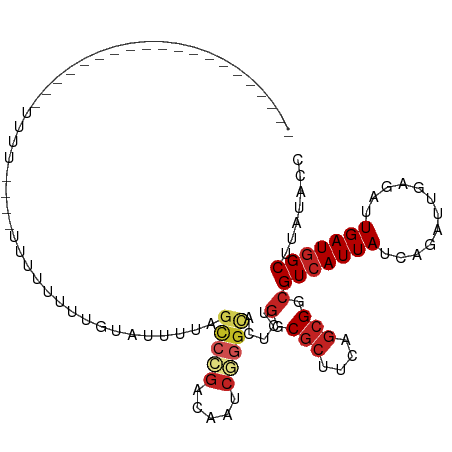
| Mean single sequence MFE | -24.08 |
| Consensus MFE | -13.51 |
| Energy contribution | -14.25 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.947571 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
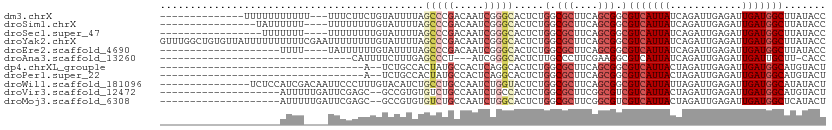
>dm3.chrX 16068315 94 - 22422827 --------------UUUUUUUUUUU---UUUCUUCUGUAUUUUAGCCCGACAAUCGGGCACUCUGGCGCUUCAGCGGCGUCAUUAUCAGAUUGAGAUUGAUGGCUUAUACC --------------.......((..---((((.((((.......(((((.....)))))....(((((((.....)))))))....))))..))))..))........... ( -25.30, z-score = -2.56, R) >droSim1.chrX 12409645 91 - 17042790 ----------------UAUUUUUU----UUUUUUUUGUAUUUUAGCCCGACAAUCGGGCACUCUGGCGCUUCAGCGGCGUCAUUAUCAGAUUGAGAUUGAUGGCUUAUACC ----------------((((..((----((..(((.(((.....(((((.....)))))....(((((((.....))))))).))).)))..))))..))))......... ( -23.40, z-score = -2.12, R) >droSec1.super_47 125822 90 - 216421 -----------------UUUUUUU----UUUUUUUUGUAUUUUAGCCCGACAAUCGGGCACUCUGGCGCUUCAGCGGCGUCAUUAUCAGAUUGAGAUUGAUGGCUUAUACC -----------------.......----........((((....(((((.....))))).....(.(((....))).)(((((((((.......)).)))))))..)))). ( -23.00, z-score = -2.04, R) >droYak2.chrX 10217022 111 - 21770863 GUUUGGCUGUGUUAUUUUUUUUUUCGAAUUUUUUUUGUAUUUUAGCCCGACAAUCGGGCACUCUGGCGCUUCAGCGGCGUCAUUAUCAGAUUGAGAUUGAUGGCUUAUACC ((..((((..((((.((((..(((.((.................(((((.....)))))....(((((((.....)))))))...)))))..)))).))))))))...)). ( -27.50, z-score = -1.40, R) >droEre2.scaffold_4690 7736168 87 + 18748788 --------------------UUUU----UAUUUUUUGUAUUUUAGCCCGACAAUCGGGCACUCUGGCGCUUCAGCGGCGUCAUUAUCAGAUUGAGAUUGAUGGCUUAUACC --------------------....----........((((....(((((.....))))).....(.(((....))).)(((((((((.......)).)))))))..)))). ( -23.00, z-score = -1.92, R) >droAna3.scaffold_13260 1918117 75 - 2125536 --------------------------------CAUUUUCUUUGAGCCCU---AUCGGGCACUCUUGCCCUUCGAAGGCGUCAUUAUCAGAUUGAGAUUGAUUGCUU-CACC --------------------------------......(((((((....---...(((((....))))))))))))((((((...((.....))...))).)))..-.... ( -16.81, z-score = -0.59, R) >dp4.chrXL_group1e 9696687 75 - 12523060 ----------------------------------A--UCUGCCACUAUGCCACUCAGGCACUCUGGCGCUUCAGCGGCGUCAUUACUAGAUUGAGAUUGAUGGCAUGUACU ----------------------------------.--..(((((...((((.....))))((((((((((.....)))))))..........))).....)))))...... ( -21.00, z-score = -0.53, R) >droPer1.super_22 1179118 75 + 1688296 ----------------------------------A--UCUGCCACUAUGCCACUCAGGCACUCUGGCGCUUCAGCGGCGUCAUUACUAGAUUGAGAUUGAUGGCAUGUACU ----------------------------------.--..(((((...((((.....))))((((((((((.....)))))))..........))).....)))))...... ( -21.00, z-score = -0.53, R) >droWil1.scaffold_181096 6823804 96 + 12416693 ---------------UCUCCAUCGACAAUUCCCUUUGUACAUCUGCCUGCCAAUCUGGUACUCUGGCGCUUCAGCGGCGUCAUUAUUAGAUUGAGAUUGAUGGCAUAUACU ---------------...((((((((((......))))............((((((((((...(((((((.....))))))).))))))))))....))))))........ ( -26.80, z-score = -1.92, R) >droVir3.scaffold_12472 372629 89 + 763072 --------------------AUUUUUGAUUCGAGC--GCCGUGUGUCUGCCAAUCUGCCACUCUGGCGCUUCGGCGUCGUCAUUACUAGAUUGAGAUUGAUGGCAUGUACU --------------------...............--(((((..((((..(((((((......((((((....)))))).......)))))))))))..)))))....... ( -27.12, z-score = -1.09, R) >droMoj3.scaffold_6308 3087894 89 - 3356042 --------------------AUUUUUGAUUCGAGC--GCCGUGUGUCUGCCAAUCUGGCACUCUGGCGCUUCGGCGUCGUCAUUACUAGAUUGAGAUUGAUGGCUCAUACU --------------------.............(.--(((((..((((..((((((((.....((((((....))))))......))))))))))))..))))).)..... ( -30.00, z-score = -1.97, R) >consensus ____________________UUUU____UUUUUUUUGUAUUUUAGCCCGACAAUCGGGCACUCUGGCGCUUCAGCGGCGUCAUUAUCAGAUUGAGAUUGAUGGCUUAUACC ............................................(((((.....))))).....(.(((....))).)(((((((............)))))))....... (-13.51 = -14.25 + 0.75)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:48:49 2011