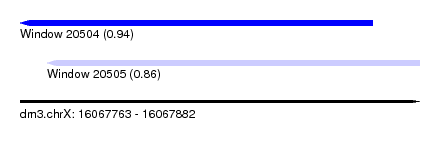
| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,067,763 – 16,067,882 |
| Length | 119 |
| Max. P | 0.935144 |

| Location | 16,067,763 – 16,067,868 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 74.38 |
| Shannon entropy | 0.47260 |
| G+C content | 0.41529 |
| Mean single sequence MFE | -24.28 |
| Consensus MFE | -17.95 |
| Energy contribution | -16.57 |
| Covariance contribution | -1.38 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.935144 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
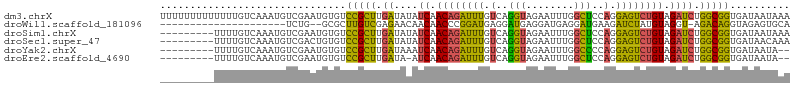
>dm3.chrX 16067763 105 - 22422827 UUUUUUUUUUUUUGUCAAAUGUCGAAUGUGUCCGCUUGAUAUAUCAACAGAUUUGUCAGGUAGAAUUUGGCUCCAGGAGUCUGUAGAUCUGGCGGUGAUAAUAAA ...........((((((...(((((((...((..(((((((.(((....))).)))))))..))))))))).((((..(((....)))))))...)))))).... ( -26.10, z-score = -1.69, R) >droWil1.scaffold_181096 6823202 81 + 12416693 ---------------------UCUG--GCGCUUGUCGAGAACAACAACCCGGAUGAGGAUGAGGAUGAGGAUGAAGAUCUAUGUAGGU-AGACAGGUAGAGUGCA ---------------------....--(((((((((....(((.((.((((........)).)).)).((((....)))).)))....-.)))).....))))). ( -13.90, z-score = 1.28, R) >droSim1.chrX 12409127 96 - 17042790 ---------UUUUGUCAAAUGUCGAAUGUGUCCGCUUGAUAUAUCAACAGAUUUGUCAGGUAGAAUUUGGCUCCAGGAGUCUGUAGAUCUGGCGGUGAUAAUAAA ---------..((((((...(((((((...((..(((((((.(((....))).)))))))..))))))))).((((..(((....)))))))...)))))).... ( -26.10, z-score = -1.86, R) >droSec1.super_47 125313 96 - 216421 ---------UUUUGUCAAAUGUCGACUGUGUCCGCUUGAUAUAUCAACAGAUUUGUCAGGUAGAAUUUGGCUCCAGGAGUCUGUAGAUCUGGCGGUGAUAACAAA ---------.(((((....((((.(((((.((..(((((((.(((....))).)))))))..))....(((((...)))))..........)))))))))))))) ( -24.80, z-score = -0.88, R) >droYak2.chrX 10216489 94 - 21770863 ---------UUUUGUCAAAUGUCGAAUGUGUCCGCUUGAUAAAUCAACAGAUUUGUCAGGUAGAAUUUGGCCCCAGGAGUCUGUAGAUCUGGCGGUGAUAAUA-- ---------..(((((......(((((...((..(((((((((((....)))))))))))..)))))))(((((((..(((....))))))).))))))))..-- ( -30.10, z-score = -3.26, R) >droEre2.scaffold_4690 7735642 93 + 18748788 ---------UUUUGUCAAAUGUCGAAUGUGUCCGCUUGAUA-AUCAACAGAUUUGUCAGGUAGAAUUUGGCUCCAGGAGUCUGUAGAUCUGGCGGUGAUAAUA-- ---------..((((((...(((((((...((..(((((((-(((....)).))))))))..))))))))).((((..(((....)))))))...))))))..-- ( -24.70, z-score = -1.43, R) >consensus _________UUUUGUCAAAUGUCGAAUGUGUCCGCUUGAUAUAUCAACAGAUUUGUCAGGUAGAAUUUGGCUCCAGGAGUCUGUAGAUCUGGCGGUGAUAAUAAA ...............................(((((......(((.((((((((.(..(((........)))..).)))))))).)))..))))).......... (-17.95 = -16.57 + -1.38)

| Location | 16,067,771 – 16,067,882 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 94.79 |
| Shannon entropy | 0.09040 |
| G+C content | 0.45634 |
| Mean single sequence MFE | -31.12 |
| Consensus MFE | -28.14 |
| Energy contribution | -28.34 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.856855 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
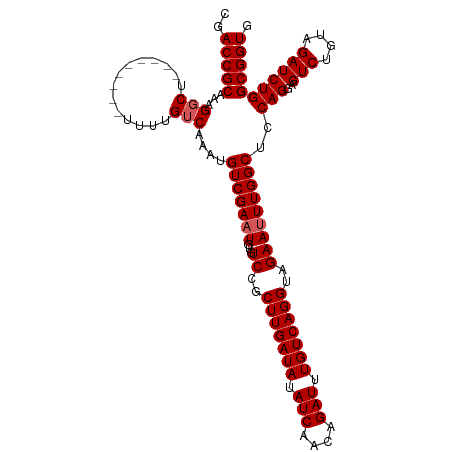
>dm3.chrX 16067771 111 - 22422827 CGACCGCAAAGUCUUUUUUUUUUUUUUGUCAAAUGUCGAAUGUGUCCGCUUGAUAUAUCAACAGAUUUGUCAGGUAGAAUUUGGCUCCAGGAGUCUGUAGAUCUGGCGGUG ..(((((...((((........((((((......(((((((...((..(((((((.(((....))).)))))))..)))))))))..)))))).....))))...))))). ( -29.72, z-score = -1.69, R) >droSim1.chrX 12409135 102 - 17042790 CGACCGCAAAGGCU---------UUUUGUCAAAUGUCGAAUGUGUCCGCUUGAUAUAUCAACAGAUUUGUCAGGUAGAAUUUGGCUCCAGGAGUCUGUAGAUCUGGCGGUG ..(((((...(((.---------....)))....(((((((...((..(((((((.(((....))).)))))))..)))))))))..(((..(((....))))))))))). ( -30.80, z-score = -1.74, R) >droSec1.super_47 125321 102 - 216421 CGACCGCAAAGGCU---------UUUUGUCAAAUGUCGACUGUGUCCGCUUGAUAUAUCAACAGAUUUGUCAGGUAGAAUUUGGCUCCAGGAGUCUGUAGAUCUGGCGGUG ..(((((..(((((---------(((.(((((((.(((((...))).((((((((.(((....))).)))))))).)))))))))...)))))))).........))))). ( -32.00, z-score = -1.87, R) >droYak2.chrX 10216495 102 - 21770863 CGACCGCAAAGGCU---------UUUUGUCAAAUGUCGAAUGUGUCCGCUUGAUAAAUCAACAGAUUUGUCAGGUAGAAUUUGGCCCCAGGAGUCUGUAGAUCUGGCGGUG ..(((((...(((.---------....)))....(((((((...((..(((((((((((....)))))))))))..)))))))))..(((..(((....))))))))))). ( -33.70, z-score = -2.69, R) >droEre2.scaffold_4690 7735648 101 + 18748788 CGACCGCAAAGGCU---------UUUUGUCAAAUGUCGAAUGUGUCCGCUUGAUA-AUCAACAGAUUUGUCAGGUAGAAUUUGGCUCCAGGAGUCUGUAGAUCUGGCGGUG ..(((((...(((.---------....)))....(((((((...((..(((((((-(((....)).))))))))..)))))))))..(((..(((....))))))))))). ( -29.40, z-score = -1.35, R) >consensus CGACCGCAAAGGCU_________UUUUGUCAAAUGUCGAAUGUGUCCGCUUGAUAUAUCAACAGAUUUGUCAGGUAGAAUUUGGCUCCAGGAGUCUGUAGAUCUGGCGGUG ..(((((...........................(((((((...((..(((((((.(((....))).)))))))..)))))))))..(((..(((....))))))))))). (-28.14 = -28.34 + 0.20)

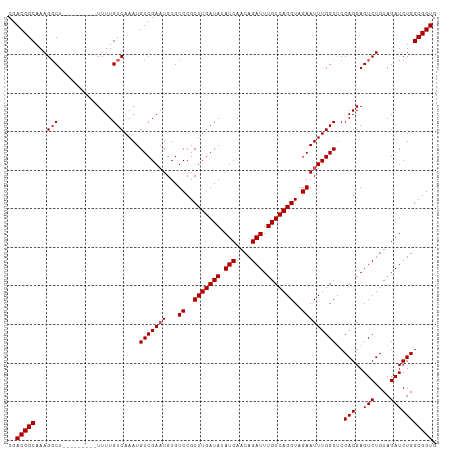
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:48:48 2011