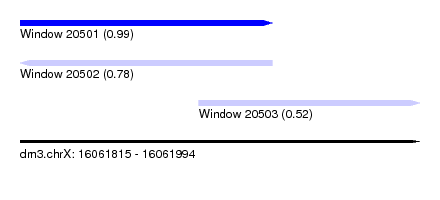
| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,061,815 – 16,061,994 |
| Length | 179 |
| Max. P | 0.989410 |

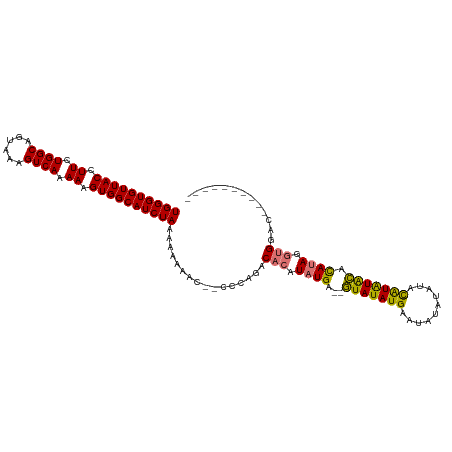
| Location | 16,061,815 – 16,061,928 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 79.34 |
| Shannon entropy | 0.32313 |
| G+C content | 0.37604 |
| Mean single sequence MFE | -30.35 |
| Consensus MFE | -21.21 |
| Energy contribution | -20.53 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.37 |
| SVM RNA-class probability | 0.989410 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 16061815 113 + 22422827 UGGGUGUUACCUUCUGGCAGUAAAGUCAAAAAGUGGCAUCUAAAAAAAAAAACCCAGACACAUAUGA--GUAUAUGAAUAUAUAUGUAUAUACACAUAUGCGCACAUAGGUGGAC ((((((((((.((.((((......)))).)).))))))))))................(((.((((.--(((((((..(((((....)))))..)))))))...)))).)))... ( -29.70, z-score = -2.83, R) >droSim1.chrX 12402699 101 + 17042790 UGGGUGUUACCUUCUGGCAGUAAAGUCAAAAAGUGGCAUCUAAAAAAAAC--CCCAGACACAUAUGA--GUAUAUGAAUAUAUAUACAUAUGCACAUAGGUGGAC---------- ((((((((((.((.((((......)))).)).))))))))))........--......(((.((((.--(((((((.((....)).))))))).)))).)))...---------- ( -28.00, z-score = -2.83, R) >droSec1.super_47 119360 99 + 216421 UGGGUGUUACCUUCUGGCAGUAAAGUCAAAAAGUGGCAUCUAAAAAAACC--CCCAGACACAUAUGA--GUAUAUGAAUAUAU--ACAUAUGCACAUAGGUGGAC---------- ((((((((((.((.((((......)))).)).))))))))))........--......(((.((((.--(((((((.......--.))))))).)))).)))...---------- ( -28.60, z-score = -2.78, R) >droEre2.scaffold_4690 7730316 110 - 18748788 UGGGUGUUACCUUCUGGCAGUAAAGUCAGAAAGUGGCAUCUAAAAAG-----CCCAGACACAUAUGACUAUAUAUGUACGCACAUAUGUGUAUAUACACAUGUACAUUGGUGGAC ((((((((((.(((((((......))))))).)))))))))).....-----......(((..(((((....((((((((((....)))))))))).....)).)))..)))... ( -35.10, z-score = -2.49, R) >consensus UGGGUGUUACCUUCUGGCAGUAAAGUCAAAAAGUGGCAUCUAAAAAAAAC__CCCAGACACAUAUGA__GUAUAUGAAUAUAUAUACAUAUACACAUAGGUGGAC__________ ((((((((((.((.((((......)))).)).))))))))))................(((.((((...(((((((..........))))))).)))).)))............. (-21.21 = -20.53 + -0.69)

| Location | 16,061,815 – 16,061,928 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 79.34 |
| Shannon entropy | 0.32313 |
| G+C content | 0.37604 |
| Mean single sequence MFE | -26.80 |
| Consensus MFE | -18.36 |
| Energy contribution | -17.30 |
| Covariance contribution | -1.06 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.784425 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 16061815 113 - 22422827 GUCCACCUAUGUGCGCAUAUGUGUAUAUACAUAUAUAUUCAUAUAC--UCAUAUGUGUCUGGGUUUUUUUUUUUAGAUGCCACUUUUUGACUUUACUGCCAGAAGGUAACACCCA ....(((((.(.(((((((((.((((((............))))))--.))))))))))))))).................((((((((.(......).))))))))........ ( -26.00, z-score = -1.91, R) >droSim1.chrX 12402699 101 - 17042790 ----------GUCCACCUAUGUGCAUAUGUAUAUAUAUUCAUAUAC--UCAUAUGUGUCUGGG--GUUUUUUUUAGAUGCCACUUUUUGACUUUACUGCCAGAAGGUAACACCCA ----------.....((((.(..((((((..(((((....))))).--.))))))..).))))--................((((((((.(......).))))))))........ ( -22.80, z-score = -1.20, R) >droSec1.super_47 119360 99 - 216421 ----------GUCCACCUAUGUGCAUAUGU--AUAUAUUCAUAUAC--UCAUAUGUGUCUGGG--GGUUUUUUUAGAUGCCACUUUUUGACUUUACUGCCAGAAGGUAACACCCA ----------((..((((....(((((((.--.((((....)))).--.))))))).(((((.--(((....(((((........)))))....))).))))))))).))..... ( -24.70, z-score = -1.36, R) >droEre2.scaffold_4690 7730316 110 + 18748788 GUCCACCAAUGUACAUGUGUAUAUACACAUAUGUGCGUACAUAUAUAGUCAUAUGUGUCUGGG-----CUUUUUAGAUGCCACUUUCUGACUUUACUGCCAGAAGGUAACACCCA (((((...((((((((((((......))))))))))))((((((((....)))))))).))))-----)............((((((((.(......).))))))))........ ( -33.70, z-score = -2.62, R) >consensus __________GUCCACCUAUGUGCAUAUAUAUAUAUAUUCAUAUAC__UCAUAUGUGUCUGGG__GUUUUUUUUAGAUGCCACUUUUUGACUUUACUGCCAGAAGGUAACACCCA .................((((((.(((((....))))).)))))).........(((((((((........))))))))).((((((((.(......).))))))))........ (-18.36 = -17.30 + -1.06)

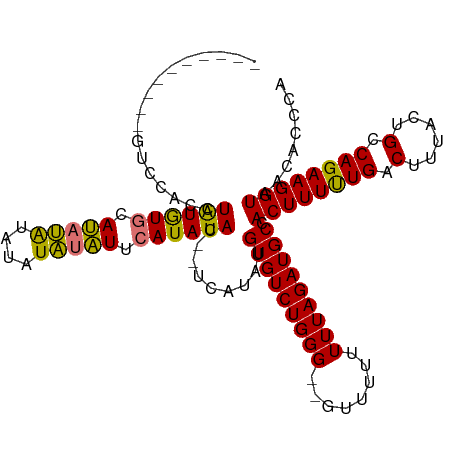
| Location | 16,061,895 – 16,061,994 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 83.84 |
| Shannon entropy | 0.24912 |
| G+C content | 0.44068 |
| Mean single sequence MFE | -19.70 |
| Consensus MFE | -16.15 |
| Energy contribution | -16.65 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.517923 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
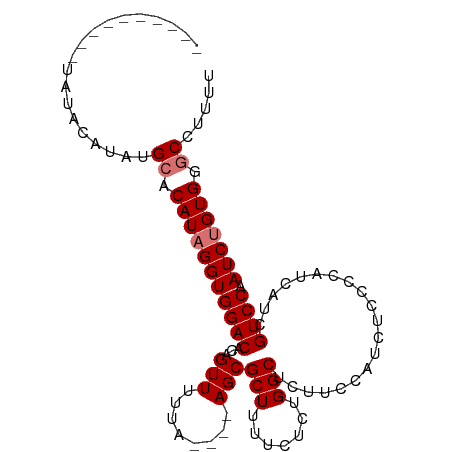
>dm3.chrX 16061895 99 + 22422827 UAUGUAUAUACACAUAUGCGCACAUAGGUGGACACAGUUUUUA----AGCGCUUUUCUCUGGCUCUUCCAUCUCGCCAUCAUCGUCCAAUCUGUGGGCCUUUU ...((((((....))))))((.(((((((((((...(((....----))).........((((...........)))).....)))).))))))).))..... ( -24.10, z-score = -2.02, R) >droSim1.chrX 12402777 89 + 17042790 ----------UAUACAUAUGCACAUAGGUGGACACAGUUUUUA----AGCGCUUUUCUCUGGCUCUACCAUCUCCCCAUCAUCGUCCAAUCUGUGGGCCUUUU ----------.........((.(((((((((((...(((....----)))(((.......)))....................)))).))))))).))..... ( -18.50, z-score = -1.05, R) >droSec1.super_47 119436 89 + 216421 ----------UAUACAUAUGCACAUAGGUGGACACAGUUUUUA----AGCGCUUUUCUCUGGCUCCUCCAUCUCCCCAUCAUCGUCCAAUCUGUGGGCCUUUU ----------.........((.(((((((((((...(((....----)))(((.......)))....................)))).))))))).))..... ( -18.50, z-score = -1.05, R) >droEre2.scaffold_4690 7730397 98 - 18748788 ----UGUGUAUAUACACAUGUACAUUGGUGGACACAGUUUUUAUUUAAGCGCUUUUCUCUGGCUCUUUCAUCUCCCCAUCAUCGUCCAAUCUGUGGCCUUUU- ----(((((....))))).(..(((.(((((((...((((......))))(((.......)))....................)))).))).)))..)....- ( -17.70, z-score = -0.87, R) >consensus __________UAUACAUAUGCACAUAGGUGGACACAGUUUUUA____AGCGCUUUUCUCUGGCUCUUCCAUCUCCCCAUCAUCGUCCAAUCUGUGGGCCUUUU ...................((.(((((((((((...(((........)))(((.......)))....................)))).))))))).))..... (-16.15 = -16.65 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:48:46 2011