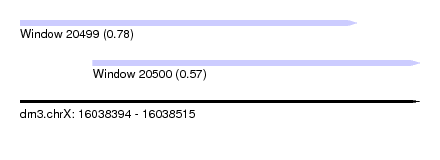
| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,038,394 – 16,038,515 |
| Length | 121 |
| Max. P | 0.777141 |

| Location | 16,038,394 – 16,038,496 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 78.38 |
| Shannon entropy | 0.28499 |
| G+C content | 0.58573 |
| Mean single sequence MFE | -42.10 |
| Consensus MFE | -22.74 |
| Energy contribution | -23.30 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.777141 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
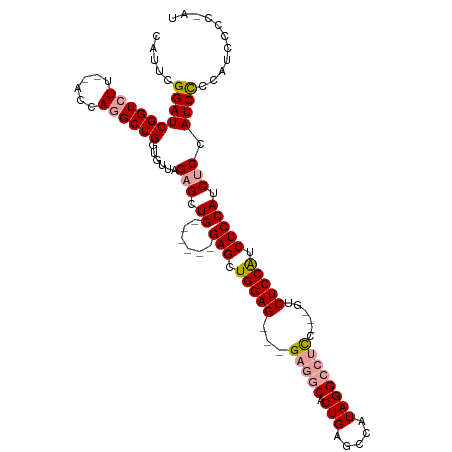
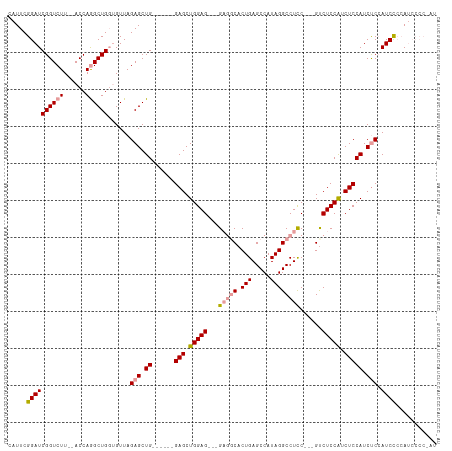
>dm3.chrX 16038394 102 + 22422827 CAUUCGGAUCGGUCUU--ACCAGGCUGGUGUUAGAGCUG------GAGCUGGAG---GAGGCACUGAGCCAUAGGCCUCC---GUCUCCAUCUCCAUCUCCAUCCGAAUCCCCCAU .((((((((((((((.--...))))))......(((.((------(((.(((((---(((((.(((.....)))))))))---...)))).))))).))).))))))))....... ( -42.90, z-score = -2.54, R) >droSec1.super_47 95821 103 + 216421 CAUUCGGAUCGGUCUUGCACCAGGCUGGUGUUAGAGCUG------GAGCUGGAG---G---CACUGAGCCAUAGGCCUCCUCCGUCUCCAUCUCCAUCCCAAUCCCCAUCCCC-AU .....((((.(((((.(((((.....))))).)))..((------(((.(((((---(---(...(((((...)).)))....))))))).)))))..)).))))........-.. ( -37.90, z-score = -1.71, R) >droEre2.scaffold_4690 7706422 104 - 18748788 CAUUCGGAUCGGUCUU--ACCAAGCUGCAGUUAGAGCUGCACCUGGAGCUGGAGCUGGAGGCACUGAGCCAUAGGCCUUC---GCCUCCGUCUCCAUCUCCAUCUCCAG------- .....(((..((....--.)).((.((((((....)))))).))((((.(((((.(((((((...(.(((...))))...---))))))).))))).))))...)))..------- ( -45.50, z-score = -3.26, R) >consensus CAUUCGGAUCGGUCUU__ACCAGGCUGGUGUUAGAGCUG______GAGCUGGAG___GAGGCACUGAGCCAUAGGCCUCC___GUCUCCAUCUCCAUCUCCAUCCCCAUCCCC_AU .....((((.(((.....))).((((........)))).......(((.(((((...(((((...(((((...)).)))....)))))...))))).))).))))........... (-22.74 = -23.30 + 0.56)

| Location | 16,038,416 – 16,038,515 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 69.97 |
| Shannon entropy | 0.39614 |
| G+C content | 0.60045 |
| Mean single sequence MFE | -33.70 |
| Consensus MFE | -19.68 |
| Energy contribution | -19.79 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.566407 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
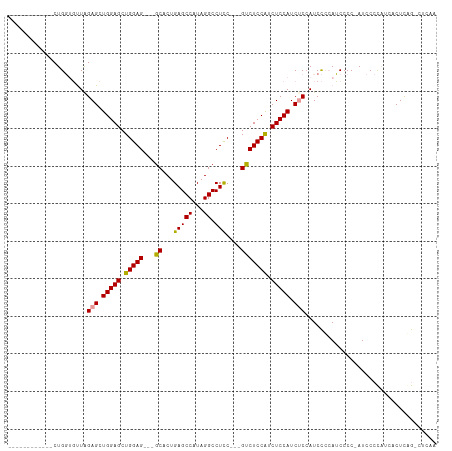
>dm3.chrX 16038416 99 + 22422827 ------------CUGGUGUUAGAGCUGGAGCUGGAGGAGGCACUGAGCCAUAGGCCUCC---GUCUCCAUCUCCAUCUCCAUCCGAAUCCCCCAUCCCCAUCACUCCGCCUCAA ------------..((((...(((.(((((.((((((((((.(((.....)))))))))---...)))).))))).))).....((........))..........)))).... ( -31.50, z-score = -1.07, R) >droSec1.super_47 95845 98 + 216421 ------------CUGGUGUUAGAGCUGGAGCUGGAG---GCACUGAGCCAUAGGCCUCCUCCGUCUCCAUCUCCAUCCCAAUCCCCAUCCCC-AUCCCCAUCUCUUGGACUCAA ------------.(((.(...(((.(((((.(((((---(....(.(((...)))).)))))).))))).)))...))))............-....(((.....)))...... ( -28.90, z-score = -0.83, R) >droEre2.scaffold_4690 7706444 101 - 18748788 CUGCAGUUAGAGCUGCACCUGGAGCUGGAGCUGGAG---GCACUGAGCCAUAGGCCUUC---GCCUCCGUCUCCAUCUCCAUCUCCAGCCCC-AUCCCUCCGCCUCAA------ .((((((....))))))..(((.(((((((.(((((---(....(((.(..((((....---))))..).)))..)))))).))))))).))-)..............------ ( -40.70, z-score = -2.70, R) >consensus ____________CUGGUGUUAGAGCUGGAGCUGGAG___GCACUGAGCCAUAGGCCUCC___GUCUCCAUCUCCAUCUCCAUCCCCAUCCCC_AUCCCCAUCACUCAG_CUCAA .....................(((.(((((.(((((........(.(((...))))........))))).))))).)))................................... (-19.68 = -19.79 + 0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:48:44 2011