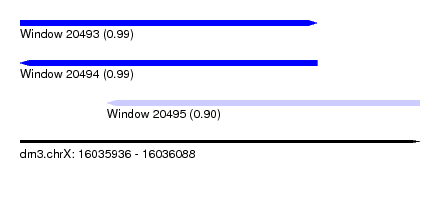
| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,035,936 – 16,036,088 |
| Length | 152 |
| Max. P | 0.993471 |

| Location | 16,035,936 – 16,036,049 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.51 |
| Shannon entropy | 0.07821 |
| G+C content | 0.43701 |
| Mean single sequence MFE | -33.10 |
| Consensus MFE | -33.40 |
| Energy contribution | -32.92 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.43 |
| Structure conservation index | 1.01 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.62 |
| SVM RNA-class probability | 0.993471 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
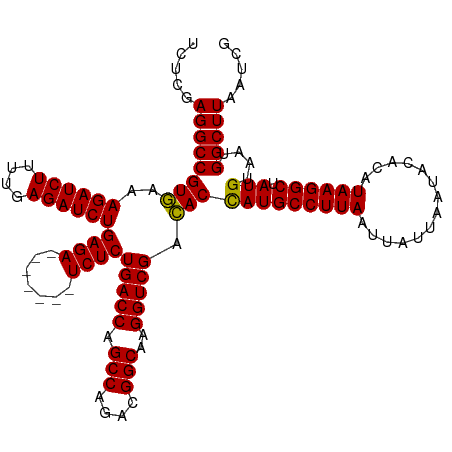
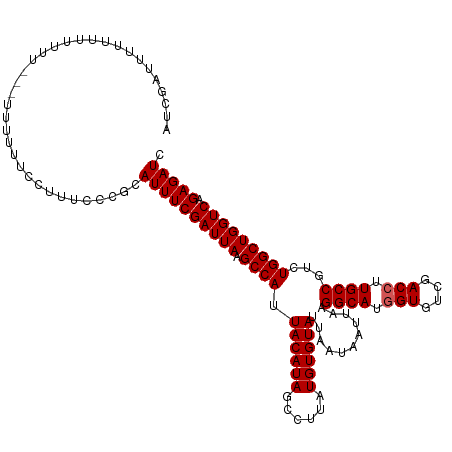
>dm3.chrX 16035936 113 + 22422827 UCUCGAGGCCGUUAAAGAUCUUUUGAGAUCUGAGA-------UCUCUGACCAGCCAGAUGGCAAGGUCGAAACCAUGCCUUAAUUAUUAAUACACAUAAGGCUAUGUAAUGGCUUAAUCG .....(((((((((..((((((..((((((...))-------))))......(((....))))))))).....(((((((((..............)))))).))))))))))))..... ( -33.94, z-score = -2.98, R) >droSim1.chrX 12373714 113 + 17042790 UCUCGAGGCCGUGAAAGAUCUUUUGAGAUCUGAGA-------UCUCUGACCAGCCAGACGGCAAGGUCGACACCAUGCCUUAAUUAUUAAUACACAUAAGGCUAUGUAAUGGCUUAAUCG .....((((((((...((((((..((((((...))-------))))......(((....)))))))))..)))(((((((((..............)))))).)))....)))))..... ( -32.14, z-score = -2.14, R) >droSec1.super_47 93383 113 + 216421 UCUCGAGGCCGUGAAAGAUCUUUUGAGAUCUGAGA-------UCUCUGACCAGCCAGACGGCAAGGUCGACACCAUGCCUUAAUUAUUAAUACACAUAAGGCUAUGUAAUGGCUUAAUCG .....((((((((...((((((..((((((...))-------))))......(((....)))))))))..)))(((((((((..............)))))).)))....)))))..... ( -32.14, z-score = -2.14, R) >droYak2.chrX 10183492 120 + 21770863 UCUCGAGGCCGUGAAAGAUCUUUUGAGAUCUGAGAACUGAGAUCUCUGACCAGCCAGACGGCAAGGUCGACACCAUGCCUUAAUUAUUAAUACACAUAAGGCUAUGUAAUGGCUUAAUCG .....((((((((...((((((..(((((((.(....).)))))))......(((....)))))))))..)))(((((((((..............)))))).)))....)))))..... ( -35.94, z-score = -2.78, R) >droEre2.scaffold_4690 7704082 113 - 18748788 UCUCGAGGCCGUGAAAGAUCUUUUGAGAUCUGAGA-------UCUCUGACCAGCCACCCGGCAAGGUCGACACUAUGCCUUAAUUAUUAAUACACAUAAGGCUAUGUAAUGGCUUAAUCG .....((((((((...((((((..((((((...))-------))))......(((....)))))))))..)))...((((((..............))))))........)))))..... ( -31.34, z-score = -2.10, R) >consensus UCUCGAGGCCGUGAAAGAUCUUUUGAGAUCUGAGA_______UCUCUGACCAGCCAGACGGCAAGGUCGACACCAUGCCUUAAUUAUUAAUACACAUAAGGCUAUGUAAUGGCUUAAUCG .....((((((((..((((((....))))))((((.......))))(((((.(((....)))..))))).)))(((((((((..............)))))).)))....)))))..... (-33.40 = -32.92 + -0.48)

| Location | 16,035,936 – 16,036,049 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.51 |
| Shannon entropy | 0.07821 |
| G+C content | 0.43701 |
| Mean single sequence MFE | -35.18 |
| Consensus MFE | -35.32 |
| Energy contribution | -35.52 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.48 |
| Structure conservation index | 1.00 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.59 |
| SVM RNA-class probability | 0.993121 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 16035936 113 - 22422827 CGAUUAAGCCAUUACAUAGCCUUAUGUGUAUUAAUAAUUAAGGCAUGGUUUCGACCUUGCCAUCUGGCUGGUCAGAGA-------UCUCAGAUCUCAAAAGAUCUUUAACGGCCUCGAGA (((....(((........((((((..(((....)))..))))))..((((((((((..(((....))).)))).))))-------))..((((((....)))))).....))).)))... ( -34.80, z-score = -2.99, R) >droSim1.chrX 12373714 113 - 17042790 CGAUUAAGCCAUUACAUAGCCUUAUGUGUAUUAAUAAUUAAGGCAUGGUGUCGACCUUGCCGUCUGGCUGGUCAGAGA-------UCUCAGAUCUCAAAAGAUCUUUCACGGCCUCGAGA (((....(((....(((.((((((..(((....)))..)))))))))(((..((((..(((....))).)))).((((-------((...))))))...........)))))).)))... ( -35.00, z-score = -2.43, R) >droSec1.super_47 93383 113 - 216421 CGAUUAAGCCAUUACAUAGCCUUAUGUGUAUUAAUAAUUAAGGCAUGGUGUCGACCUUGCCGUCUGGCUGGUCAGAGA-------UCUCAGAUCUCAAAAGAUCUUUCACGGCCUCGAGA (((....(((....(((.((((((..(((....)))..)))))))))(((..((((..(((....))).)))).((((-------((...))))))...........)))))).)))... ( -35.00, z-score = -2.43, R) >droYak2.chrX 10183492 120 - 21770863 CGAUUAAGCCAUUACAUAGCCUUAUGUGUAUUAAUAAUUAAGGCAUGGUGUCGACCUUGCCGUCUGGCUGGUCAGAGAUCUCAGUUCUCAGAUCUCAAAAGAUCUUUCACGGCCUCGAGA (((....(((....(((.((((((..(((....)))..)))))))))(((..((((..(((....))).)))).(((((((........)))))))...........)))))).)))... ( -37.30, z-score = -2.64, R) >droEre2.scaffold_4690 7704082 113 + 18748788 CGAUUAAGCCAUUACAUAGCCUUAUGUGUAUUAAUAAUUAAGGCAUAGUGUCGACCUUGCCGGGUGGCUGGUCAGAGA-------UCUCAGAUCUCAAAAGAUCUUUCACGGCCUCGAGA (((....(((........((((((..(((....)))..))))))...(((..((((..(((....))).)))).((((-------((...))))))...........)))))).)))... ( -33.80, z-score = -1.94, R) >consensus CGAUUAAGCCAUUACAUAGCCUUAUGUGUAUUAAUAAUUAAGGCAUGGUGUCGACCUUGCCGUCUGGCUGGUCAGAGA_______UCUCAGAUCUCAAAAGAUCUUUCACGGCCUCGAGA (((...(((((.((((((......))))))...........((((.(((....))).))))...)))))((((.((((.......))))((((((....)))))).....)))))))... (-35.32 = -35.52 + 0.20)

| Location | 16,035,969 – 16,036,088 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 93.33 |
| Shannon entropy | 0.12356 |
| G+C content | 0.39306 |
| Mean single sequence MFE | -26.08 |
| Consensus MFE | -25.10 |
| Energy contribution | -25.30 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.96 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.899992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
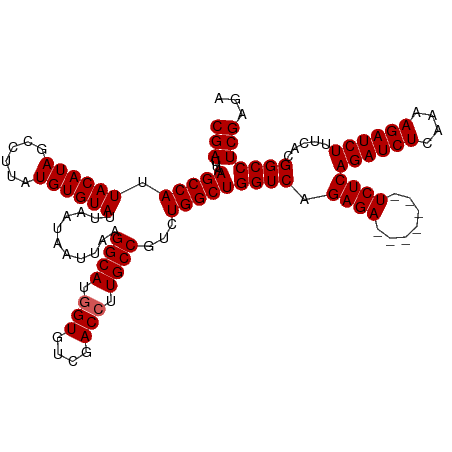
>dm3.chrX 16035969 119 - 22422827 AUCGAUUUUUUUUGUUCGUUUUUUUCCUUUCCCGCAUUUCGAUUAAGCCAUUACAUAGCCUUAUGUGUAUUAAUAAUUAAGGCAUGGUUUCGACCUUGCCAUCUGGCUGGUCAGAGAUC ..(((..........))).................((((((((((.((((.((((((......))))))...........((((.(((....))).))))...))))))))).))))). ( -26.00, z-score = -1.79, R) >droSim1.chrX 12373747 118 - 17042790 AUCGAUUUUUUUCUUUCG-UUUUUUCCUUUCCCGCAUUUCGAUUAAGCCAUUACAUAGCCUUAUGUGUAUUAAUAAUUAAGGCAUGGUGUCGACCUUGCCGUCUGGCUGGUCAGAGAUC ..(((..........)))-................((((((((((.((((.((((((......))))))...........((((.(((....))).))))...))))))))).))))). ( -26.20, z-score = -1.54, R) >droSec1.super_47 93416 115 - 216421 AUCGAUUUUUUUUCG----UUUUUUCCUUUCCCGCAUUUCGAUUAAGCCAUUACAUAGCCUUAUGUGUAUUAAUAAUUAAGGCAUGGUGUCGACCUUGCCGUCUGGCUGGUCAGAGAUC ..(((.......)))----................((((((((((.((((.((((((......))))))...........((((.(((....))).))))...))))))))).))))). ( -26.80, z-score = -1.78, R) >droYak2.chrX 10183532 119 - 21770863 AUCGAUUUUUUUUUUUGUUUUUUUUCCUUUCCCGCAUUUCGAUUAAGCCAUUACAUAGCCUUAUGUGUAUUAAUAAUUAAGGCAUGGUGUCGACCUUGCCGUCUGGCUGGUCAGAGAUC ...................................((((((((((.((((.((((((......))))))...........((((.(((....))).))))...))))))))).))))). ( -26.10, z-score = -1.72, R) >droEre2.scaffold_4690 7704115 112 + 18748788 AUCGAUUUUUCG-------UUUUUUCCUUUCCCGCAUUUCGAUUAAGCCAUUACAUAGCCUUAUGUGUAUUAAUAAUUAAGGCAUAGUGUCGACCUUGCCGGGUGGCUGGUCAGAGAUC ..(((....)))-------................((((((((((.(((((((((((......))))))...........((((..((....))..))))..)))))))))).))))). ( -25.30, z-score = -0.84, R) >consensus AUCGAUUUUUUUUUUU___UUUUUUCCUUUCCCGCAUUUCGAUUAAGCCAUUACAUAGCCUUAUGUGUAUUAAUAAUUAAGGCAUGGUGUCGACCUUGCCGUCUGGCUGGUCAGAGAUC ...................................((((((((((.((((.((((((......))))))...........((((.(((....))).))))...))))))))).))))). (-25.10 = -25.30 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:48:40 2011