| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,031,262 – 16,031,359 |
| Length | 97 |
| Max. P | 0.500000 |

| Location | 16,031,262 – 16,031,359 |
|---|---|
| Length | 97 |
| Sequences | 12 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 77.32 |
| Shannon entropy | 0.50784 |
| G+C content | 0.50848 |
| Mean single sequence MFE | -27.15 |
| Consensus MFE | -15.09 |
| Energy contribution | -15.83 |
| Covariance contribution | 0.74 |
| Combinations/Pair | 1.14 |
| Mean z-score | -0.98 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chrX 16031262 97 + 22422827 GCCUGUAAAAAGCAAUUCACUGUAAUUGCUUUGUGACUUAUGGCCCGGCCCAACUUGCGCGUUCAUCGCU-UUACCGAUUGCUAGUCGGCAGGUAGCU (((.((..(((((((((......)))))))))...))....)))..(((...(((((((((.....))).-....((((.....)))))))))).))) ( -28.60, z-score = -1.00, R) >droSim1.chrX 12369154 97 + 17042790 GCCUGUAAAAAGCAAUUCACUGUAAUUGCUUUGUGACUUAUGGCCCGGCCCAACUUGCGCGUUCAUCGCU-UUACCGAUUGCUAGUCGGCAGGUAGCU (((.((..(((((((((......)))))))))...))....)))..(((...(((((((((.....))).-....((((.....)))))))))).))) ( -28.60, z-score = -1.00, R) >droSec1.super_47 88794 97 + 216421 GCCUGUAAAAAGCAAUUCACUGUAAUUGCUUUGUGACUUAUGGCCCGACCCAACUUGCGCGUUCAUCGCU-UUACCGAUUGCUAGUCGGCAGGUAGCU (((.((..(((((((((......)))))))))...))....)))........(((((((((.....))).-....((((.....)))))))))).... ( -27.20, z-score = -1.30, R) >droYak2.chrX 10179005 97 + 21770863 GCCUGUAAAAAGCAAUUCACUGUAAUUGCUUUGUGACUUAUGGCCCGGCCCAACUUGCGCGUUCAUCGCU-UUACCGAUUGCUAGUCGGCAGGUAGCU (((.((..(((((((((......)))))))))...))....)))..(((...(((((((((.....))).-....((((.....)))))))))).))) ( -28.60, z-score = -1.00, R) >droEre2.scaffold_4690 7699515 97 - 18748788 GCCUGUAAAAAGCAAUUCACUGUAAUUGCUUUGUGACUUAUGGCCCGGCCCAACUUGCGCGUUCAUCGCU-UUACCGAUUGCUAGUCGGCAGGUAGCU (((.((..(((((((((......)))))))))...))....)))..(((...(((((((((.....))).-....((((.....)))))))))).))) ( -28.60, z-score = -1.00, R) >droAna3.scaffold_13260 1806580 82 + 2125536 ACUUGUGGAAAGCAAUUCAAUGUAAUUGCUUUGUGACUUAUAGCCCGACCUAACUUGCGA---------U-GUGAAUAAUGCUAGUCGGCAG------ ...(((.((.(((((((((.....(((((...((..................))..))))---------)-.)))))..))))..)).))).------ ( -15.27, z-score = 0.19, R) >dp4.chrXL_group1e 9650169 98 + 12523060 GCCUGUAGAAAGCAAUUCACUGUAAUUGCUUUGUGACUUAUGGCCCGGCCCAACUUGCGCGUCCAUCGUUCGUGGCCACAGUCGUGGGGCAGGUAGCC (((((((.(((((((((......))))))))).)..((((((((..((((..((..(((.......)))..))))))...)))))))))))))).... ( -33.50, z-score = -0.80, R) >droPer1.super_22 1132381 98 - 1688296 GCCUGUAGAAAGCAAUUCACUGUAAUUGCUUUGUGACUUAUGGCCCGGCCCAACUUGCCCGUCCAUCGUUCGUGGCCACAGUCGCGGGGCAGGUAGCC ((((((..(((((((((......)))))))))((((((..(((((((((.......)))((.....))...).))))).))))))...)))))).... ( -35.40, z-score = -1.51, R) >droWil1.scaffold_181096 6770053 75 - 12416693 GCCUGUAGAAAGCAAUUCACUGUAAUUGCUUUGUGACUUAUGGCCCAAAAGCAGCCAUCCAGCCAUC--UAGUCAGC--------------------- ........(((((((((......))))))))).(((((.(((((.................))))).--.)))))..--------------------- ( -17.83, z-score = -1.01, R) >droVir3.scaffold_12472 324742 89 - 763072 GCCUGUAGAAAGCAAUUCACUGUAAUUGCUUUGUGACUUAUGGCCCGGCCCAACUUGUGCGCUCGCCGAUCGU-----CAGUCG----GCAGGUAGCC ((((((..(((((((((......))))))))).(((((.((((..((((...............)))).))))-----.)))))----)))))).... ( -27.96, z-score = -0.85, R) >droMoj3.scaffold_6308 3036178 89 + 3356042 GCCUGUAGAAAGCAAUUCACUGUAAUUGCUUUGUGACUUAUGGCCCGGCCCAACUUGUGCGCUCGCCGAUCGU-----CAGUCG----GCAGGUAGCC ((((((..(((((((((......))))))))).(((((.((((..((((...............)))).))))-----.)))))----)))))).... ( -27.96, z-score = -0.85, R) >droGri2.scaffold_15203 4521471 88 + 11997470 GCCUGUAGAAAGCAAUUCACUGUAAUUGCUUUGUGACUUAUGGCCCGACCCAACUUCUGCGCUCGCCGAUCGU-----CAGUCG----GCAGGUAAC- (((((((((((((((((......)))))))).........(((......)))...)))))....((((((...-----..))))----))))))...- ( -26.30, z-score = -1.69, R) >consensus GCCUGUAGAAAGCAAUUCACUGUAAUUGCUUUGUGACUUAUGGCCCGGCCCAACUUGCGCGUUCAUCGCU_GUACCGACUGCUAGUCGGCAGGUAGCU ((((((..(((((((((......)))))))))(((.....(((......))).....)))............................)))))).... (-15.09 = -15.83 + 0.74)

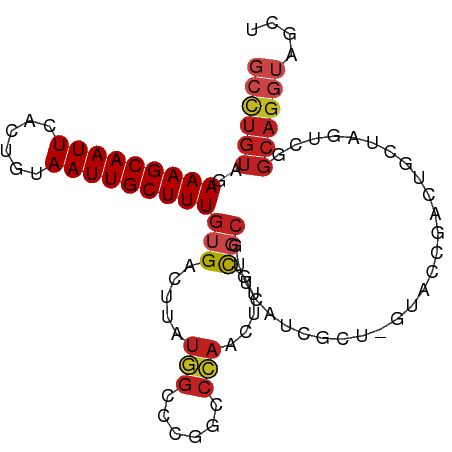

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:48:38 2011