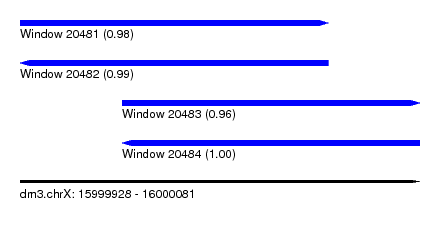
| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,999,928 – 16,000,081 |
| Length | 153 |
| Max. P | 0.998373 |

| Location | 15,999,928 – 16,000,046 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 93.84 |
| Shannon entropy | 0.08488 |
| G+C content | 0.33424 |
| Mean single sequence MFE | -31.57 |
| Consensus MFE | -28.54 |
| Energy contribution | -29.43 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.978405 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15999928 118 + 22422827 AAGCGC-GCGUAGUAGCUAUUUUUGGAGACUAAACUACCGCCAUCUGUUGGUGUUUAUUCAAAACAGACGUAAAAAAGUAUGUUACACAAAAUAAAUUUUUUGUGUAGUUACUAUAUUU ......-..(((((((((..(((((((...(((((.((((.(....).))))))))))))))))..((((((......))))))((((((((......))))))))))))))))).... ( -29.00, z-score = -2.22, R) >droSim1.chrX 12341025 119 + 17042790 AAGCGCUGCGUAGUAGCCAUUUUUGGAGACUUAAUUGUCGCCAUCUGUUGGUGUUUAUUCACAACAGACGUGUAAAAGUAUGUUACACAAAAUAAAUUUUUUGUGUAGUUACUAUAUUU .........((((((((.((((((((.(((......))).)).(((((((.((......))))))))).....))))))....(((((((((......))))))))))))))))).... ( -33.10, z-score = -3.02, R) >droSec1.super_47 57761 119 + 216421 AAGCGCUGCGUAGUAGCCAUUUUUGGAGGCUUAAUUGUCGCCAUCUGUUGGUGUUUAUUCACAACAGACGUGAAAAAGUAUGUUACACAAAAUAAUUUUUUUGUGUAGUUACUAUAUUU .........((((((((.((((((((.(((......))).)).(((((((.((......))))))))).....))))))....(((((((((......))))))))))))))))).... ( -32.60, z-score = -2.62, R) >consensus AAGCGCUGCGUAGUAGCCAUUUUUGGAGACUUAAUUGUCGCCAUCUGUUGGUGUUUAUUCACAACAGACGUGAAAAAGUAUGUUACACAAAAUAAAUUUUUUGUGUAGUUACUAUAUUU .........((((((((.((((((((.(((......))).)).(((((((.((......))))))))).....))))))....(((((((((......))))))))))))))))).... (-28.54 = -29.43 + 0.89)


| Location | 15,999,928 – 16,000,046 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 93.84 |
| Shannon entropy | 0.08488 |
| G+C content | 0.33424 |
| Mean single sequence MFE | -28.37 |
| Consensus MFE | -24.38 |
| Energy contribution | -24.82 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.34 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.56 |
| SVM RNA-class probability | 0.992715 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15999928 118 - 22422827 AAAUAUAGUAACUACACAAAAAAUUUAUUUUGUGUAACAUACUUUUUUACGUCUGUUUUGAAUAAACACCAACAGAUGGCGGUAGUUUAGUCUCCAAAAAUAGCUACUACGC-GCGCUU ......((((..(((((((((......)))))))))...))))......((((((((.((......))..))))))))((((((((..(((...........)))))))).)-)).... ( -26.80, z-score = -2.66, R) >droSim1.chrX 12341025 119 - 17042790 AAAUAUAGUAACUACACAAAAAAUUUAUUUUGUGUAACAUACUUUUACACGUCUGUUGUGAAUAAACACCAACAGAUGGCGACAAUUAAGUCUCCAAAAAUGGCUACUACGCAGCGCUU ......((((..(((((((((......)))))))))...))))......(((((((((((......)).)))))))))(((.......(((..(((....)))..)))......))).. ( -28.72, z-score = -3.51, R) >droSec1.super_47 57761 119 - 216421 AAAUAUAGUAACUACACAAAAAAAUUAUUUUGUGUAACAUACUUUUUCACGUCUGUUGUGAAUAAACACCAACAGAUGGCGACAAUUAAGCCUCCAAAAAUGGCUACUACGCAGCGCUU ......((((..(((((((((......)))))))))...))))......(((((((((((......)).)))))))))(((.......((((.........)))).........))).. ( -29.59, z-score = -3.84, R) >consensus AAAUAUAGUAACUACACAAAAAAUUUAUUUUGUGUAACAUACUUUUUCACGUCUGUUGUGAAUAAACACCAACAGAUGGCGACAAUUAAGUCUCCAAAAAUGGCUACUACGCAGCGCUU ......((((..(((((((((......)))))))))...))))......(((((((((((......)).)))))))))(((.......((((.........)))).........))).. (-24.38 = -24.82 + 0.45)

| Location | 15,999,967 – 16,000,081 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 85.84 |
| Shannon entropy | 0.19112 |
| G+C content | 0.28421 |
| Mean single sequence MFE | -24.77 |
| Consensus MFE | -22.06 |
| Energy contribution | -21.40 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.962319 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15999967 114 + 22422827 CCAUCUGUUGGUGUUUAUUCAAAACAGACGUAAAAAAGUAUGUUACACAAAAUAAAUUUUUUGUGUAGUUACUAUAUUUUUAGGGUUGACUAGCGUUGUGCUAUUUUUCAGUAA ((.((((((..((......)).)))))).....(((((((((((((((((((......))))))))))....))))))))).))(..((.((((.....)))).))..)..... ( -24.40, z-score = -1.67, R) >droSim1.chrX 12341065 104 + 17042790 CCAUCUGUUGGUGUUUAUUCACAACAGACGUGUAAAAGUAUGUUACACAAAAUAAAUUUUUUGUGUAGUUACUAUAUUUUUAGGGUUAACUGUGAUUGUUAUAA---------- ((.(((((((.((......)))))))))....((((((((((((((((((((......))))))))))....))))))))))))..((((.......))))...---------- ( -25.60, z-score = -2.48, R) >droSec1.super_47 57801 102 + 216421 CCAUCUGUUGGUGUUUAUUCACAACAGACGUGAAAAAGUAUGUUACACAAAAUAAUUUUUUUGUGUAGUUACUAUAUUUUUAGGGUUAACUGUGAUUGUUAU------------ ((.(((((((.((......))))))))).....(((((((((((((((((((......))))))))))....))))))))).))..((((.......)))).------------ ( -24.30, z-score = -2.19, R) >consensus CCAUCUGUUGGUGUUUAUUCACAACAGACGUGAAAAAGUAUGUUACACAAAAUAAAUUUUUUGUGUAGUUACUAUAUUUUUAGGGUUAACUGUGAUUGUUAUA___________ ((.(((((((.((......))))))))).....(((((((((((((((((((......))))))))))....))))))))).))......(((((...)))))........... (-22.06 = -21.40 + -0.66)

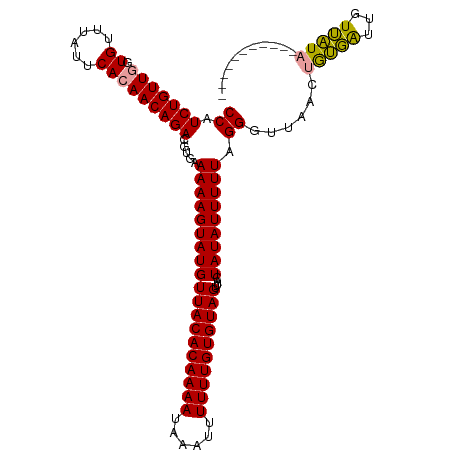
| Location | 15,999,967 – 16,000,081 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 85.84 |
| Shannon entropy | 0.19112 |
| G+C content | 0.28421 |
| Mean single sequence MFE | -22.83 |
| Consensus MFE | -20.03 |
| Energy contribution | -20.37 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.69 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.34 |
| SVM RNA-class probability | 0.998373 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15999967 114 - 22422827 UUACUGAAAAAUAGCACAACGCUAGUCAACCCUAAAAAUAUAGUAACUACACAAAAAAUUUAUUUUGUGUAACAUACUUUUUUACGUCUGUUUUGAAUAAACACCAACAGAUGG ....(((....((((.....)))).)))....((((((...((((..(((((((((......)))))))))...))))))))))((((((((.((......))..)))))))). ( -23.70, z-score = -3.81, R) >droSim1.chrX 12341065 104 - 17042790 ----------UUAUAACAAUCACAGUUAACCCUAAAAAUAUAGUAACUACACAAAAAAUUUAUUUUGUGUAACAUACUUUUACACGUCUGUUGUGAAUAAACACCAACAGAUGG ----------...((((.......)))).............((((..(((((((((......)))))))))...))))......(((((((((((......)).))))))))). ( -22.40, z-score = -3.51, R) >droSec1.super_47 57801 102 - 216421 ------------AUAACAAUCACAGUUAACCCUAAAAAUAUAGUAACUACACAAAAAAAUUAUUUUGUGUAACAUACUUUUUCACGUCUGUUGUGAAUAAACACCAACAGAUGG ------------.((((.......)))).............((((..(((((((((......)))))))))...))))......(((((((((((......)).))))))))). ( -22.40, z-score = -3.74, R) >consensus ___________UAUAACAAUCACAGUUAACCCUAAAAAUAUAGUAACUACACAAAAAAUUUAUUUUGUGUAACAUACUUUUUCACGUCUGUUGUGAAUAAACACCAACAGAUGG .........................................((((..(((((((((......)))))))))...))))......(((((((((((......)).))))))))). (-20.03 = -20.37 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:48:31 2011