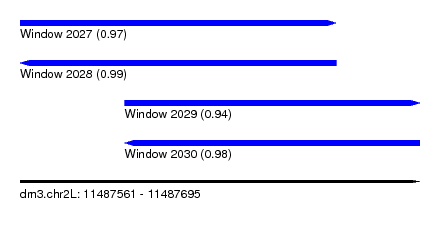
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,487,561 – 11,487,695 |
| Length | 134 |
| Max. P | 0.993012 |

| Location | 11,487,561 – 11,487,667 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 95.09 |
| Shannon entropy | 0.08614 |
| G+C content | 0.50082 |
| Mean single sequence MFE | -40.60 |
| Consensus MFE | -34.64 |
| Energy contribution | -34.52 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.967141 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
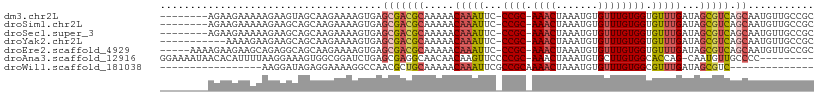
>dm3.chr2L 11487561 106 + 23011544 CAAAGUGGGUGGUGGCCAAAGGGUGAACC-----ACCCAAGCGGCAACAUUGCUGACGCUAUCAAACACCACAAACACAUUUAGUUUGCGGGAAUUUGUUUUUGCGUCGCU .....((((((((.(((....)))..)))-----)))))(((((((....))))(((((...((((..((.(((((.......))))).))...)))).....)))))))) ( -41.40, z-score = -3.36, R) >droSim1.chr2L 11301884 106 + 22036055 CAUAGUGGGUGGUGGCCAAAGGGUGACCC-----ACCCAAGCGGCAACAUUGCUGACGCUAUCAAACACCACAAACACAUUUAGUUUGCGGGAAUUUGUUUUUGCGUCGCU .....((((((((.(((....))).).))-----)))))(((((((....))))(((((...((((..((.(((((.......))))).))...)))).....)))))))) ( -40.30, z-score = -2.90, R) >droSec1.super_3 6882610 106 + 7220098 CAAAGUGGGUGGUGGCCAAAGGGUGACCC-----ACCCAAGCGGCAACAUUGCUGACGCUAUCAAACACCACAAACACAUUUAGUUUGCGGGAAUUUGUUUUUGCGUCGCU .....((((((((.(((....))).).))-----)))))(((((((....))))(((((...((((..((.(((((.......))))).))...)))).....)))))))) ( -40.30, z-score = -2.79, R) >droYak2.chr2L 7893942 111 + 22324452 GAAAGUGGGUGGUGGCCAAAGGGUGACCCGAGCCACCCAAGCGGCAACAUUGCUGACGCUAUCAAACACCACAAACACAUUUAGUUUGCGGGAAUUUGUUUUUGCGUCGCU .....((((((((.......((....))...))))))))(((((((....))))(((((...((((..((.(((((.......))))).))...)))).....)))))))) ( -41.40, z-score = -2.36, R) >droEre2.scaffold_4929 12695257 106 - 26641161 GUGGGUGGGUGGUGGUUAAAGGGUGACCC-----ACCCAAGCGGCAACAUUGCUGACGCUAUCAAACACCACAAACACAUUUAGUUUGCGGGAAUUUGUUUUUGCGUCGCU .(((((((((.(...........).))))-----)))))(((((((....))))(((((...((((..((.(((((.......))))).))...)))).....)))))))) ( -39.60, z-score = -2.28, R) >consensus CAAAGUGGGUGGUGGCCAAAGGGUGACCC_____ACCCAAGCGGCAACAUUGCUGACGCUAUCAAACACCACAAACACAUUUAGUUUGCGGGAAUUUGUUUUUGCGUCGCU .....((((((((.(((....))).)))......)))))(((((((....))))(((((...((((..((.(((((.......))))).))...)))).....)))))))) (-34.64 = -34.52 + -0.12)

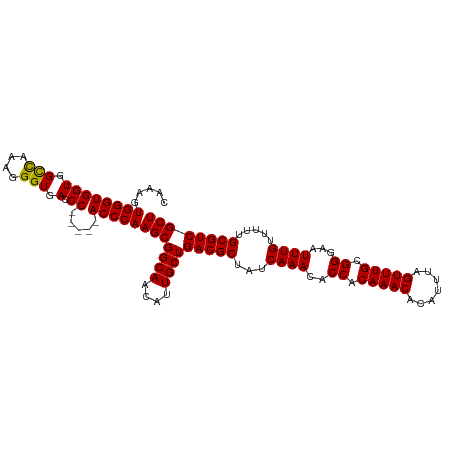
| Location | 11,487,561 – 11,487,667 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 95.09 |
| Shannon entropy | 0.08614 |
| G+C content | 0.50082 |
| Mean single sequence MFE | -40.29 |
| Consensus MFE | -34.98 |
| Energy contribution | -35.22 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.30 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.58 |
| SVM RNA-class probability | 0.993012 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 11487561 106 - 23011544 AGCGACGCAAAAACAAAUUCCCGCAAACUAAAUGUGUUUGUGGUGUUUGAUAGCGUCAGCAAUGUUGCCGCUUGGGU-----GGUUCACCCUUUGGCCACCACCCACUUUG ((((((((.....(((((..((((((((.......)))))))).)))))...))))).(((....))).)))(((((-----(((...((....))..))))))))..... ( -39.20, z-score = -3.35, R) >droSim1.chr2L 11301884 106 - 22036055 AGCGACGCAAAAACAAAUUCCCGCAAACUAAAUGUGUUUGUGGUGUUUGAUAGCGUCAGCAAUGUUGCCGCUUGGGU-----GGGUCACCCUUUGGCCACCACCCACUAUG ((((((((.....(((((..((((((((.......)))))))).)))))...))))).(((....))).)))(((((-----((((((.....))))..)))))))..... ( -39.40, z-score = -3.01, R) >droSec1.super_3 6882610 106 - 7220098 AGCGACGCAAAAACAAAUUCCCGCAAACUAAAUGUGUUUGUGGUGUUUGAUAGCGUCAGCAAUGUUGCCGCUUGGGU-----GGGUCACCCUUUGGCCACCACCCACUUUG ((((((((.....(((((..((((((((.......)))))))).)))))...))))).(((....))).)))(((((-----((((((.....))))..)))))))..... ( -39.40, z-score = -2.92, R) >droYak2.chr2L 7893942 111 - 22324452 AGCGACGCAAAAACAAAUUCCCGCAAACUAAAUGUGUUUGUGGUGUUUGAUAGCGUCAGCAAUGUUGCCGCUUGGGUGGCUCGGGUCACCCUUUGGCCACCACCCACUUUC ((((((((.....(((((..((((((((.......)))))))).)))))...))))).(((....))).)))(((((((....(((((.....))))).)))))))..... ( -43.30, z-score = -3.32, R) >droEre2.scaffold_4929 12695257 106 + 26641161 AGCGACGCAAAAACAAAUUCCCGCAAACUAAAUGUGUUUGUGGUGUUUGAUAGCGUCAGCAAUGUUGCCGCUUGGGU-----GGGUCACCCUUUAACCACCACCCACCCAC ((((((((.....(((((..((((((((.......)))))))).)))))...))))).(((....))).)))(((((-----((((...............))))))))). ( -40.16, z-score = -3.89, R) >consensus AGCGACGCAAAAACAAAUUCCCGCAAACUAAAUGUGUUUGUGGUGUUUGAUAGCGUCAGCAAUGUUGCCGCUUGGGU_____GGGUCACCCUUUGGCCACCACCCACUUUG ((((((((.....(((((..((((((((.......)))))))).)))))...))))).(((....))).)))(((((......(((((.....)))))...)))))..... (-34.98 = -35.22 + 0.24)

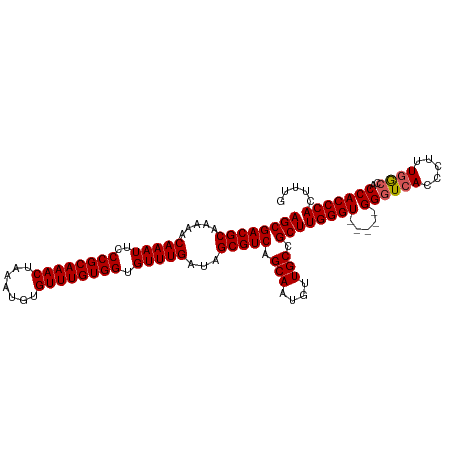
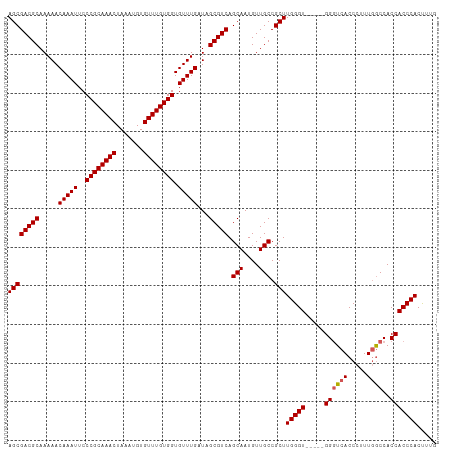
| Location | 11,487,596 – 11,487,695 |
|---|---|
| Length | 99 |
| Sequences | 7 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 73.22 |
| Shannon entropy | 0.50776 |
| G+C content | 0.43520 |
| Mean single sequence MFE | -24.67 |
| Consensus MFE | -8.59 |
| Energy contribution | -8.84 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.944174 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 11487596 99 + 23011544 GCGGCAACAUUGCUGACGCUAUCAAACACCACAAACACAUUUAGUUU-GCGG-GAAUUUGUUUUUGCGUCGCUCACUUUUCUUGCUACUUCUUUUUCUUCU-------- ((((((....))))(((((...((((..((.(((((.......))))-).))-...)))).....))))))).............................-------- ( -22.40, z-score = -2.91, R) >droSim1.chr2L 11301919 99 + 22036055 GCGGCAACAUUGCUGACGCUAUCAAACACCACAAACACAUUUAGUUU-GCGG-GAAUUUGUUUUUGCGUCGCUCACUUUUCUUGCUGCUUCUUUUUCUUCU-------- (((((((....((.(((((...((((..((.(((((.......))))-).))-...)))).....))))))).........))))))).............-------- ( -27.12, z-score = -3.91, R) >droSec1.super_3 6882645 99 + 7220098 GCGGCAACAUUGCUGACGCUAUCAAACACCACAAACACAUUUAGUUU-GCGG-GAAUUUGUUUUUGCGUCGCUCACUUUUCUUGCUGCUUCUUUUUCUUCU-------- (((((((....((.(((((...((((..((.(((((.......))))-).))-...)))).....))))))).........))))))).............-------- ( -27.12, z-score = -3.91, R) >droYak2.chr2L 7893982 96 + 22324452 GCGGCAACAUUGCUGACGCUAUCAAACACCACAAACACAUUUAGUUU-GCGG-GAAUUUGUUUUUGCGUCGCUCACUUUUCUUGCUGCUUCUUCUUUU----------- (((((((....((.(((((...((((..((.(((((.......))))-).))-...)))).....))))))).........)))))))..........----------- ( -27.12, z-score = -3.76, R) >droEre2.scaffold_4929 12695292 102 - 26641161 GCGGCAACAUUGCUGACGCUAUCAAACACCACAAACACAUUUAGUUU-GCGG-GAAUUUGUUUUUGCGUCGCUCACUUUUCUUGCUGCCUCUGCUUCUUCUUUU----- (((((((....((.(((((...((((..((.(((((.......))))-).))-...)))).....))))))).........)))))))................----- ( -26.72, z-score = -2.99, R) >droAna3.scaffold_12916 12187683 98 - 16180835 ---------GGGGCAACAUUG-CUGGUGCCACAAGCACAUUUAGUUU-GCGGGGAACUUGUUGUUGCCUCGCUCAGAUCCGCCACUUUCCUUAAAAUGUGUUAUUUUCC ---------(((((((((..(-(.(((.((.(((((.......))))-).))...))).)))))))))))((........))(((............)))......... ( -25.10, z-score = -0.06, R) >droWil1.scaffold_181038 383564 78 - 637489 --------------GACGCUAUCAAACGCCACAAACACAUUUAGUUUUGCGGCGAAUUUGUUUUUGCAGCGUUGGCCUUUUCCUCUAUCCUU----------------- --------------((((((......((((.((((.((.....)))))).)))).....((....))))))))...................----------------- ( -17.10, z-score = -1.52, R) >consensus GCGGCAACAUUGCUGACGCUAUCAAACACCACAAACACAUUUAGUUU_GCGG_GAAUUUGUUUUUGCGUCGCUCACUUUUCUUGCUGCUUCUUUUUCUUCU________ ..............(((((...((((..((.(((((.......)))).).))....)))).....)))))....................................... ( -8.59 = -8.84 + 0.25)
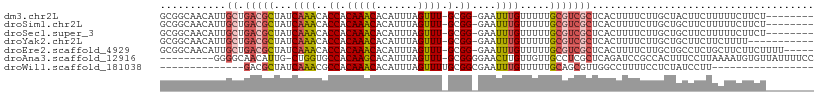
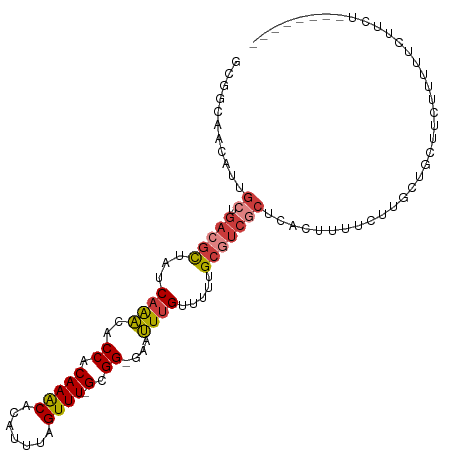
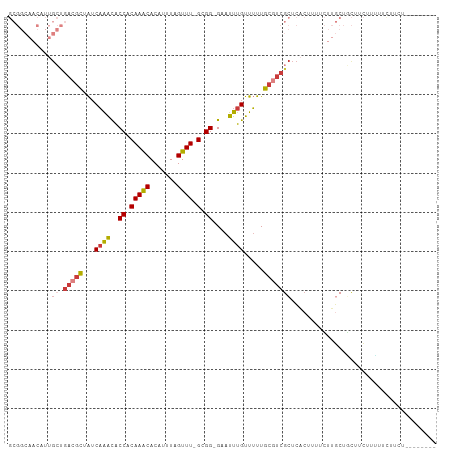
| Location | 11,487,596 – 11,487,695 |
|---|---|
| Length | 99 |
| Sequences | 7 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 73.22 |
| Shannon entropy | 0.50776 |
| G+C content | 0.43520 |
| Mean single sequence MFE | -26.77 |
| Consensus MFE | -13.38 |
| Energy contribution | -13.53 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.979713 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 11487596 99 - 23011544 --------AGAAGAAAAAGAAGUAGCAAGAAAAGUGAGCGACGCAAAAACAAAUUC-CCGC-AAACUAAAUGUGUUUGUGGUGUUUGAUAGCGUCAGCAAUGUUGCCGC --------.............((((((......(....)(((((.....(((((..-((((-((((.......)))))))).)))))...))))).....))))))... ( -25.50, z-score = -2.39, R) >droSim1.chr2L 11301919 99 - 22036055 --------AGAAGAAAAAGAAGCAGCAAGAAAAGUGAGCGACGCAAAAACAAAUUC-CCGC-AAACUAAAUGUGUUUGUGGUGUUUGAUAGCGUCAGCAAUGUUGCCGC --------.............((((((......(....)(((((.....(((((..-((((-((((.......)))))))).)))))...))))).....))))))... ( -27.50, z-score = -2.60, R) >droSec1.super_3 6882645 99 - 7220098 --------AGAAGAAAAAGAAGCAGCAAGAAAAGUGAGCGACGCAAAAACAAAUUC-CCGC-AAACUAAAUGUGUUUGUGGUGUUUGAUAGCGUCAGCAAUGUUGCCGC --------.............((((((......(....)(((((.....(((((..-((((-((((.......)))))))).)))))...))))).....))))))... ( -27.50, z-score = -2.60, R) >droYak2.chr2L 7893982 96 - 22324452 -----------AAAAGAAGAAGCAGCAAGAAAAGUGAGCGACGCAAAAACAAAUUC-CCGC-AAACUAAAUGUGUUUGUGGUGUUUGAUAGCGUCAGCAAUGUUGCCGC -----------..........((((((......(....)(((((.....(((((..-((((-((((.......)))))))).)))))...))))).....))))))... ( -27.50, z-score = -2.57, R) >droEre2.scaffold_4929 12695292 102 + 26641161 -----AAAAGAAGAAGCAGAGGCAGCAAGAAAAGUGAGCGACGCAAAAACAAAUUC-CCGC-AAACUAAAUGUGUUUGUGGUGUUUGAUAGCGUCAGCAAUGUUGCCGC -----...............(((((((......(....)(((((.....(((((..-((((-((((.......)))))))).)))))...))))).....))))))).. ( -31.70, z-score = -3.16, R) >droAna3.scaffold_12916 12187683 98 + 16180835 GGAAAAUAACACAUUUUAAGGAAAGUGGCGGAUCUGAGCGAGGCAACAACAAGUUCCCCGC-AAACUAAAUGUGCUUGUGGCACCAG-CAAUGUUGCCCC--------- ((...(((((((((((..((....((((.(((.((......(....)....)).)))))))-...))))))))).))))((((.((.-...)).))))))--------- ( -25.60, z-score = -0.52, R) >droWil1.scaffold_181038 383564 78 + 637489 -----------------AAGGAUAGAGGAAAAGGCCAACGCUGCAAAAACAAAUUCGCCGCAAAACUAAAUGUGUUUGUGGCGUUUGAUAGCGUC-------------- -----------------.........((......)).((((((((((........(((((((((((.....)).))))))))))))).)))))).-------------- ( -22.10, z-score = -2.01, R) >consensus ________AGAAGAAAAAGAAGCAGCAAGAAAAGUGAGCGACGCAAAAACAAAUUC_CCGC_AAACUAAAUGUGUUUGUGGUGUUUGAUAGCGUCAGCAAUGUUGCCGC .......................................(((((.....(((((...((((.((((.......)))))))).)))))...))))).............. (-13.38 = -13.53 + 0.15)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:32:52 2011