
| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,966,504 – 15,966,628 |
| Length | 124 |
| Max. P | 0.882283 |

| Location | 15,966,504 – 15,966,609 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 84.54 |
| Shannon entropy | 0.27369 |
| G+C content | 0.45256 |
| Mean single sequence MFE | -32.92 |
| Consensus MFE | -21.74 |
| Energy contribution | -20.50 |
| Covariance contribution | -1.24 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.882283 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
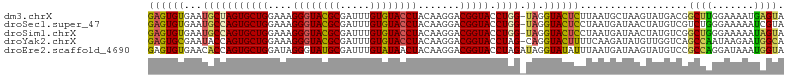
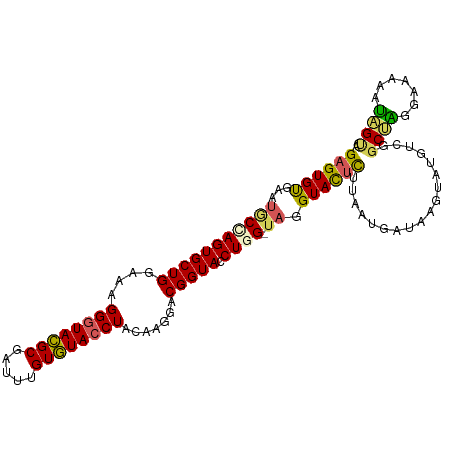
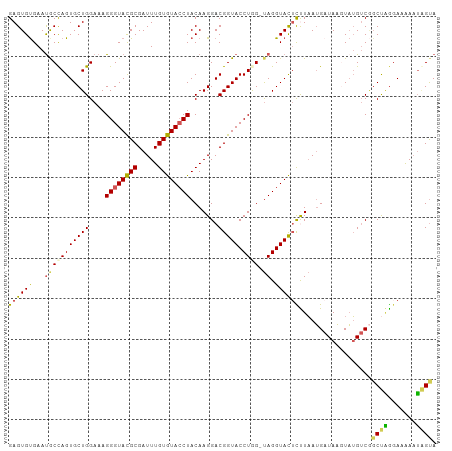
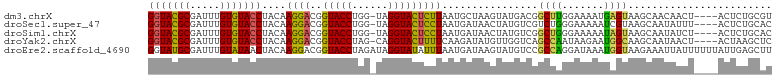
>dm3.chrX 15966504 105 + 22422827 GAGUGUGAAUGCUAGUGCUGGAAAGGGUACGCGAUUUGUGUACCUACAAGGACGGUACCUGG-UAGGUACUCUUAAUGCUAAGUAUGACGGCUUGGAAAAUGAGUA (((.(((..(((((((((((....((((((((.....)))))))).......))))).))))-))..))).)))....((((((......)))))).......... ( -32.80, z-score = -2.53, R) >droSec1.super_47 18971 105 + 216421 GAGUGUGAAUGCCAGUGCUGGAAAGGGUACGCGAUUUGUGUACCUACAAGGACGGUACCUGG-UAGGUACUCCUAAUGAUAACUAUGUCGUCUGGGAAAAAUCGUA ..(((((..(.(((....)))...)..)))))(((((..((((((((.(((......))).)-)))))))(((((((((((....)))))).))))).)))))... ( -36.10, z-score = -2.98, R) >droSim1.chrX 12326790 105 + 17042790 GAGUGUGAAUGCCAGUGCUGGAAAGGGUACGCGAUUUGUGUACCUACAAGGACGGUACCUGG-UAGGUACUCCUAAUGAUAACUAUGUCGGCUGGGAAAAAUAGUA ....(((..(((((((((((....((((((((.....)))))))).......))))).))))-))..)))(((((.(((((....)))))..)))))......... ( -35.30, z-score = -2.81, R) >droYak2.chrX 10114475 105 + 21770863 GAGUGCGAAUACCAGUGCUGGAAAGGGUACGCGAUUUGUGUACCUACAAGGACGGUACCUAG-CAGGUACUUUUCAAGAUAUGUUGGUCAGCCAAUAAGAAUGGCA ...(((.....(((....)))...((((((((.....)))))))).((.(((.((((((...-..)))))).)))......((((((....))))))....))))) ( -31.50, z-score = -1.57, R) >droEre2.scaffold_4690 7637620 106 - 18748788 GAGUGUGAACACCAGUGCUGGAUAGGGUAUGCGAUUUGUAUAACUACAAGGACGGUACCUAGAUAGGUAUAUUUAAUGAUAAGUAUGUCCGCCAGGAUAAAUGGUA ..........((((((.((((...((..((((..((((((....))))))....((((((....))))))............))))..)).)))).))...)))). ( -28.90, z-score = -1.64, R) >consensus GAGUGUGAAUGCCAGUGCUGGAAAGGGUACGCGAUUUGUGUACCUACAAGGACGGUACCUGG_UAGGUACUCUUAAUGAUAAGUAUGUCGGCUAGGAAAAAUAGUA ((((......(((.((.((.....((((((((.....))))))))...)).)))))((((....))))))))..................((((.......)))). (-21.74 = -20.50 + -1.24)
| Location | 15,966,504 – 15,966,609 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 84.54 |
| Shannon entropy | 0.27369 |
| G+C content | 0.45256 |
| Mean single sequence MFE | -23.04 |
| Consensus MFE | -14.66 |
| Energy contribution | -14.02 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.725263 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
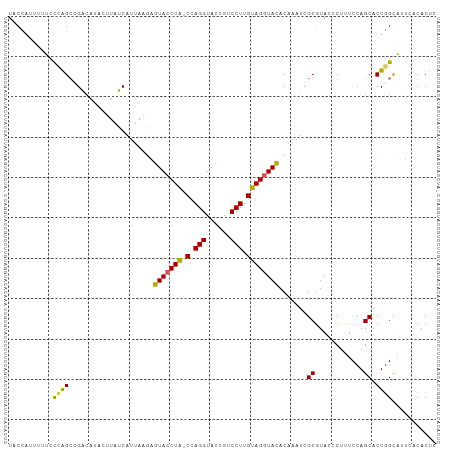
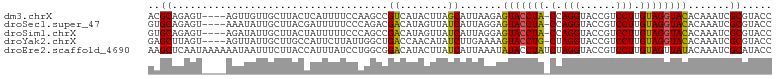
>dm3.chrX 15966504 105 - 22422827 UACUCAUUUUCCAAGCCGUCAUACUUAGCAUUAAGAGUACCUA-CCAGGUACCGUCCUUGUAGGUACACAAAUCGCGUACCCUUUCCAGCACUAGCAUUCACACUC ...........................((.......(((((((-(.(((......))).)))))))).......((............))....)).......... ( -20.40, z-score = -2.75, R) >droSec1.super_47 18971 105 - 216421 UACGAUUUUUCCCAGACGACAUAGUUAUCAUUAGGAGUACCUA-CCAGGUACCGUCCUUGUAGGUACACAAAUCGCGUACCCUUUCCAGCACUGGCAUUCACACUC ..((((((.(((..((..((...))..))....)))(((((((-(.(((......))).))))))))..))))))..........(((....)))........... ( -24.70, z-score = -2.31, R) >droSim1.chrX 12326790 105 - 17042790 UACUAUUUUUCCCAGCCGACAUAGUUAUCAUUAGGAGUACCUA-CCAGGUACCGUCCUUGUAGGUACACAAAUCGCGUACCCUUUCCAGCACUGGCAUUCACACUC ..............((((..................(((((((-(.(((......))).)))))))).......((............))..)))).......... ( -23.60, z-score = -2.28, R) >droYak2.chrX 10114475 105 - 21770863 UGCCAUUCUUAUUGGCUGACCAACAUAUCUUGAAAAGUACCUG-CUAGGUACCGUCCUUGUAGGUACACAAAUCGCGUACCCUUUCCAGCACUGGUAUUCGCACUC .((((.......))))....................(((((((-(.(((......))).)))))))).......(((........(((....)))....))).... ( -27.60, z-score = -2.32, R) >droEre2.scaffold_4690 7637620 106 + 18748788 UACCAUUUAUCCUGGCGGACAUACUUAUCAUUAAAUAUACCUAUCUAGGUACCGUCCUUGUAGUUAUACAAAUCGCAUACCCUAUCCAGCACUGGUGUUCACACUC (((((......((((.((((....(((....)))...(((((....)))))..))))(((((....)))))..............))))...)))))......... ( -18.90, z-score = -0.86, R) >consensus UACCAUUUUUCCCAGCCGACAUACUUAUCAUUAAGAGUACCUA_CCAGGUACCGUCCUUGUAGGUACACAAAUCGCGUACCCUUUCCAGCACUGGCAUUCACACUC ...........((((.....................(((((((.(.(((......))).)))))))).......((............)).))))........... (-14.66 = -14.02 + -0.64)


| Location | 15,966,529 – 15,966,628 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 79.01 |
| Shannon entropy | 0.36254 |
| G+C content | 0.41892 |
| Mean single sequence MFE | -27.98 |
| Consensus MFE | -17.86 |
| Energy contribution | -16.78 |
| Covariance contribution | -1.08 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.874157 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
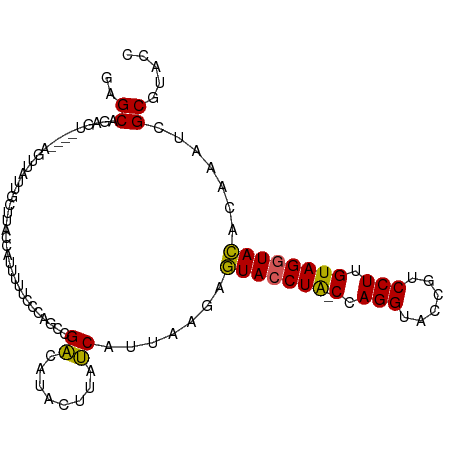
>dm3.chrX 15966529 99 + 22422827 GGUACGCGAUUUGUGUACCUACAAGGACGGUACCUGG-UAGGUACUCUUAAUGCUAAGUAUGACGGCUUGGAAAAUGAGUAAGCAACAACU----ACUCUGCGU ...(((((....(.((((((((.(((......))).)-))))))).)......((((((......)))))).....(((((.........)----))))))))) ( -29.00, z-score = -1.55, R) >droSec1.super_47 18996 99 + 216421 GGUACGCGAUUUGUGUACCUACAAGGACGGUACCUGG-UAGGUACUCCUAAUGAUAACUAUGUCGUCUGGGAAAAAUCGUAAGCAAUAUUU----ACUCUGCAC .....(((((((..((((((((.(((......))).)-)))))))(((((((((((....)))))).))))).)))))))..(((......----....))).. ( -32.40, z-score = -3.30, R) >droSim1.chrX 12326815 99 + 17042790 GGUACGCGAUUUGUGUACCUACAAGGACGGUACCUGG-UAGGUACUCCUAAUGAUAACUAUGUCGGCUGGGAAAAAUAGUAAGCAAUAUCU----ACUCUGCAC ((((.((...((((((((((((.(((......))).)-)))))))(((((.(((((....)))))..)))))...))))...))..)))).----......... ( -27.50, z-score = -1.37, R) >droYak2.chrX 10114500 99 + 21770863 GGUACGCGAUUUGUGUACCUACAAGGACGGUACCUAG-CAGGUACUUUUCAAGAUAUGUUGGUCAGCCAAUAAGAAUGGCAAGCAAUAACU----ACUAAGCUC (((((((.....))))))).....(((.((((((...-..)))))).)))......(((((.(..((((.......)))).).)))))...----......... ( -27.10, z-score = -1.66, R) >droEre2.scaffold_4690 7637645 104 - 18748788 GGUAUGCGAUUUGUAUAACUACAAGGACGGUACCUAGAUAGGUAUAUUUAAUGAUAAGUAUGUCCGCCAGGAUAAAUGGUAAGAAAUUAUUUUUUAUUGAGCUU (((.......(((((....)))))(((((((((((....))))))(((((....))))).))))))))..((((((..((((....))))..))))))...... ( -23.90, z-score = -1.78, R) >consensus GGUACGCGAUUUGUGUACCUACAAGGACGGUACCUGG_UAGGUACUCUUAAUGAUAAGUAUGUCGGCUAGGAAAAAUAGUAAGCAAUAACU____ACUCUGCAC (((((((.....)))))))....((((..((((((....))))))))))................((((.......))))........................ (-17.86 = -16.78 + -1.08)


| Location | 15,966,529 – 15,966,628 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 79.01 |
| Shannon entropy | 0.36254 |
| G+C content | 0.41892 |
| Mean single sequence MFE | -24.20 |
| Consensus MFE | -13.24 |
| Energy contribution | -12.64 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.847590 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15966529 99 - 22422827 ACGCAGAGU----AGUUGUUGCUUACUCAUUUUCCAAGCCGUCAUACUUAGCAUUAAGAGUACCUA-CCAGGUACCGUCCUUGUAGGUACACAAAUCGCGUACC ((((.((((----(((....)).)))))...............................(((((((-(.(((......))).)))))))).......))))... ( -28.40, z-score = -3.42, R) >droSec1.super_47 18996 99 - 216421 GUGCAGAGU----AAAUAUUGCUUACGAUUUUUCCCAGACGACAUAGUUAUCAUUAGGAGUACCUA-CCAGGUACCGUCCUUGUAGGUACACAAAUCGCGUACC ((((.((((----(.....))))).((((((.(((..((..((...))..))....)))(((((((-(.(((......))).))))))))..)))))).)))). ( -29.20, z-score = -3.54, R) >droSim1.chrX 12326815 99 - 17042790 GUGCAGAGU----AGAUAUUGCUUACUAUUUUUCCCAGCCGACAUAGUUAUCAUUAGGAGUACCUA-CCAGGUACCGUCCUUGUAGGUACACAAAUCGCGUACC (((((((((----((.((.....))))))))).....((.((........((.....))(((((((-(.(((......))).)))))))).....)))))))). ( -24.00, z-score = -1.59, R) >droYak2.chrX 10114500 99 - 21770863 GAGCUUAGU----AGUUAUUGCUUGCCAUUCUUAUUGGCUGACCAACAUAUCUUGAAAAGUACCUG-CUAGGUACCGUCCUUGUAGGUACACAAAUCGCGUACC .((((....----))))...((..((((.......))))....................(((((((-(.(((......))).)))))))).......))..... ( -25.10, z-score = -1.38, R) >droEre2.scaffold_4690 7637645 104 + 18748788 AAGCUCAAUAAAAAAUAAUUUCUUACCAUUUAUCCUGGCGGACAUACUUAUCAUUAAAUAUACCUAUCUAGGUACCGUCCUUGUAGUUAUACAAAUCGCAUACC ..((................................(((((......(((....)))...(((((....)))))))))).(((((....)))))...))..... ( -14.30, z-score = -1.10, R) >consensus GAGCAGAGU____AGUUAUUGCUUACCAUUUUUCCCAGCCGACAUACUUAUCAUUAAGAGUACCUA_CCAGGUACCGUCCUUGUAGGUACACAAAUCGCGUACC ..((....................................((........)).......(((((((.(.(((......))).)))))))).......))..... (-13.24 = -12.64 + -0.60)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:48:18 2011