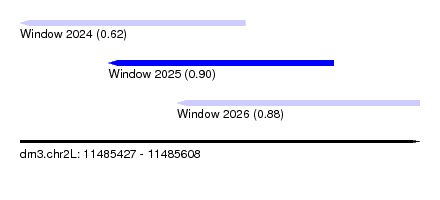
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,485,427 – 11,485,608 |
| Length | 181 |
| Max. P | 0.904922 |

| Location | 11,485,427 – 11,485,529 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 94.77 |
| Shannon entropy | 0.07202 |
| G+C content | 0.59804 |
| Mean single sequence MFE | -41.50 |
| Consensus MFE | -39.41 |
| Energy contribution | -38.87 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.95 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.621480 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 11485427 102 - 23011544 UCCAGAUGGUGGGGGGUGGUGUGAAGAAGGAGGCGGAGGUGGCGGAGGAGGAUAUCCCUGCCAAUGGUGGCCACCAUACAGAUAUCCCCCUGGUGGUGGAUA ((((......((((((((.((((.....((.(((.(...((((((.(((.....)))))))))....).))).)).))))..))))))))......)))).. ( -42.30, z-score = -1.63, R) >droSim1.chr2L 11298685 102 - 22036055 UCCAGAUGGUGGGGGGUAGUGUGAUGAAGGAGGAGGUGGUGGCGGAGGGGGAUAUCCCUGCCAAUAGUGGCCACCAUACGGAUAUCCCCCUGGUGGUGGAUA ((((......((((((((.((((.(......)..((((((.(((((((((....))))).))....)).)))))).))))..))))))))......)))).. ( -38.90, z-score = -0.45, R) >droSec1.super_3 6880457 102 - 7220098 UCCAGAUGGUGGGGGGUGGUGUGAUGAAGGAGGCGGAGGUGGCGGAGGAGGAUAUCCCUGCCAAUAGUGGCCACCAUACGGAUAUCCCCCCGGUGGUGGAUA ((((..((.(((((((..((.((.....((.(((.(...((((((.(((.....)))))))))....).))).))...)).))..))))))).)).)))).. ( -43.30, z-score = -1.47, R) >consensus UCCAGAUGGUGGGGGGUGGUGUGAUGAAGGAGGCGGAGGUGGCGGAGGAGGAUAUCCCUGCCAAUAGUGGCCACCAUACGGAUAUCCCCCUGGUGGUGGAUA ((((.((.(.((((((((.((((.....((.(((.(...((((((.(((.....)))))))))....).))).)).))))..))))))))).))..)))).. (-39.41 = -38.87 + -0.55)



| Location | 11,485,467 – 11,485,569 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 95.42 |
| Shannon entropy | 0.06302 |
| G+C content | 0.64052 |
| Mean single sequence MFE | -44.97 |
| Consensus MFE | -43.87 |
| Energy contribution | -43.43 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.98 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.904922 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 11485467 102 - 23011544 CUCCUGGGCAACCGCAAUGCGGACACCCUCCUGCUGGAGAUCCAGAUGGUGGGGGGUGGUGUGAAGAAGGAGGCGGAGGUGGCGGAGGAGGAUAUCCCUGCC (((((...((.((((...)))).((((((((..((((....)))).....))))))))...))....)))))........(((((.(((.....)))))))) ( -45.60, z-score = -1.85, R) >droSim1.chr2L 11298725 102 - 22036055 CUCCUGGGCAACCACAAUGCGGGCACCCUCCUGCUGGAGAUCCAGAUGGUGGGGGGUAGUGUGAUGAAGGAGGAGGUGGUGGCGGAGGGGGAUAUCCCUGCC (((((((....)).((...((.(((((((((..((((....)))).....))))))).)).)).))....))))).....(((((.(((.....)))))))) ( -43.10, z-score = -0.82, R) >droSec1.super_3 6880497 102 - 7220098 CUCCUGGGCAACCACAAUGCGGACACCCUCCUGCUGGAGAUCCAGAUGGUGGGGGGUGGUGUGAUGAAGGAGGCGGAGGUGGCGGAGGAGGAUAUCCCUGCC (((((((....)).((.((((..((((((((..((((....)))).....)))))))).)))).)).)))))........(((((.(((.....)))))))) ( -46.20, z-score = -2.00, R) >consensus CUCCUGGGCAACCACAAUGCGGACACCCUCCUGCUGGAGAUCCAGAUGGUGGGGGGUGGUGUGAUGAAGGAGGCGGAGGUGGCGGAGGAGGAUAUCCCUGCC (((((((....))......((.(((((((((..((((....)))).....))))))).)).))....)))))........(((((.(((.....)))))))) (-43.87 = -43.43 + -0.44)

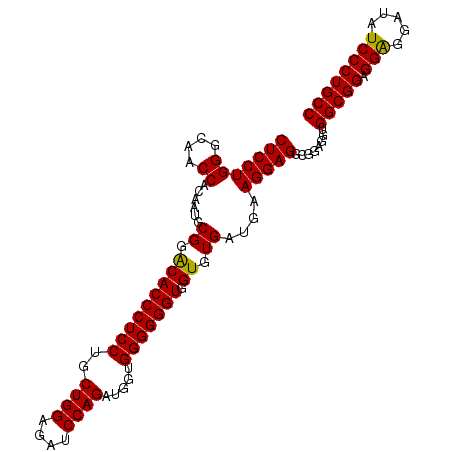
| Location | 11,485,498 – 11,485,608 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.68 |
| Shannon entropy | 0.19087 |
| G+C content | 0.59754 |
| Mean single sequence MFE | -50.70 |
| Consensus MFE | -41.66 |
| Energy contribution | -42.85 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.880713 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 11485498 110 - 23011544 -GUCUUUCGGUACCAGGAAAUCCACUUUGAGGUGGACCGCCUCCUGGGCAACCGCAAUGCGGACACCCUCCUGCUGGAGAUCCAGAUGGUGGGGGGUGGUGUGAAGAAGGA--------- -.((((((....((((((..((((((....)))))).....))))))(((.((((...)))).((((((((..((((....)))).....)))))))).)))...))))))--------- ( -50.20, z-score = -2.36, R) >droSim1.chr2L 11298756 110 - 22036055 -GUCUUUCGGUACCGGGAGAUCCAUUUUGAGGUGGACCGCCUCCUGGGCAACCACAAUGCGGGCACCCUCCUGCUGGAGAUCCAGAUGGUGGGGGGUAGUGUGAUGAAGGA--------- -.(((((((((.(((((((.((((((....))))))....)))))))...)))......((.(((((((((..((((....)))).....))))))).)).))..))))))--------- ( -47.40, z-score = -1.43, R) >droSec1.super_3 6880528 110 - 7220098 -GUCUUUCGGUACCGGGAGAUCCACUUUGAGGUGGACCGCCUCCUGGGCAACCACAAUGCGGACACCCUCCUGCUGGAGAUCCAGAUGGUGGGGGGUGGUGUGAUGAAGGA--------- -.(((((((((.(((((((.((((((....))))))....)))))))...))).....(((..((((((((..((((....)))).....)))))))).)))...))))))--------- ( -50.70, z-score = -2.13, R) >droYak2.chr2L 7891789 120 - 22324452 UGUCUUUCGGUACCAGGCGAUCCACUCUGAGGUGGACCGCCUCCUGGGCAACCACAAUGCGGACACCCUCGUGGUGGAGAUCCUGAUGGUUGCGGGUGAUGUGGUGAUGGUACCGGAGGC ...((((((((((((((...((((((.(((((((..((((....(((....)))....)))).)).))))).))))))...))).((.(..(((.....)))..).))))))))))))). ( -54.50, z-score = -2.13, R) >consensus _GUCUUUCGGUACCAGGAGAUCCACUUUGAGGUGGACCGCCUCCUGGGCAACCACAAUGCGGACACCCUCCUGCUGGAGAUCCAGAUGGUGGGGGGUGGUGUGAUGAAGGA_________ ..(((((((((.(((((((.((((((....))))))....)))))))...))).....(((..((((((((..((((....)))).....)))))))).)))...))))))......... (-41.66 = -42.85 + 1.19)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:32:49 2011