
| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,876,759 – 15,876,934 |
| Length | 175 |
| Max. P | 0.953091 |

| Location | 15,876,759 – 15,876,868 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.36 |
| Shannon entropy | 0.09399 |
| G+C content | 0.47315 |
| Mean single sequence MFE | -34.36 |
| Consensus MFE | -29.45 |
| Energy contribution | -29.45 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.872601 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15876759 109 + 22422827 CAGUUUGACCUAAAAAUGAAAAUACAAAUACCUACUGAGUCAUCGCUAUACC----ACUC--AGUAUGUGCGUGUGUCCAUGUCGGAGC-----AUGCUCCGAACGUGGGUUGCUGGCCA ((((..((((...........(((((..(((.((((((((............----))))--)))).)))..))))).((((((((((.-----...))))).))))))))))))).... ( -37.10, z-score = -3.41, R) >droEre2.scaffold_4690 7575303 115 - 18748788 CAGUUUGACCUAAAAAUGAAAAUACAAAUACCUACUGAGUCAUCGCUAUACCGAAUACUCGUAGUAUGUGCGUGUGUCCAUGUCGGAGC-----AUGCUCCGAACGCGGGUUGCUGGCCA ((((..(((((....(((...(((((..(((.((((((((..(((......)))..))))..)))).)))..))))).))).((((((.-----...))))))....))))))))).... ( -32.70, z-score = -1.51, R) >droYak2.chrX 10065550 120 + 21770863 CAGUUUGACCUAAAAAUGAAAAUACAAAUACCUACUGAGUCAUCGCUAUACCCAAUACUCGUAGUAUGUGCGUGUGUCCAUGUCGGAGCGGAGCAUGCUCCGAACGCGGGUUGCUGGCCA ((((..(((((....(((...(((((..(((.((((((((................))))..)))).)))..))))).))).((((((((.....))))))))....))))))))).... ( -31.99, z-score = -0.38, R) >droSec1.super_183 1701 109 + 33538 CAGUUUGACCUAAAAAUGAAAAUACAAAUACCUACUGAGUCAUCGCUAUACC----ACUC--AGUAUGUGUGUGUGUCCAUGUCGGAGC-----AUGCUCCGAACGCGGGUUGCUGGCCA ((((..(((((....(((...(((((.((((.((((((((............----))))--)))).)))).))))).))).((((((.-----...))))))....))))))))).... ( -35.50, z-score = -2.91, R) >droSim1.chrX 12288367 109 + 17042790 CAGUUUGACCUAAAAAUGAAAAUACAAAUACCUACUGAGUCAUCGCUAUACC----ACUC--AGUAUGUGCGUGUGUCCAUGUCGGAGC-----AUGCUCCGAACGCGGGUUGCUGGCCA ((((..(((((....(((...(((((..(((.((((((((............----))))--)))).)))..))))).))).((((((.-----...))))))....))))))))).... ( -34.50, z-score = -2.50, R) >consensus CAGUUUGACCUAAAAAUGAAAAUACAAAUACCUACUGAGUCAUCGCUAUACC____ACUC__AGUAUGUGCGUGUGUCCAUGUCGGAGC_____AUGCUCCGAACGCGGGUUGCUGGCCA ((((..(((((....(((...(((((..(((.((((((((................))))..)))).)))..))))).))).(((((((.......)))))))....))))))))).... (-29.45 = -29.45 + -0.00)


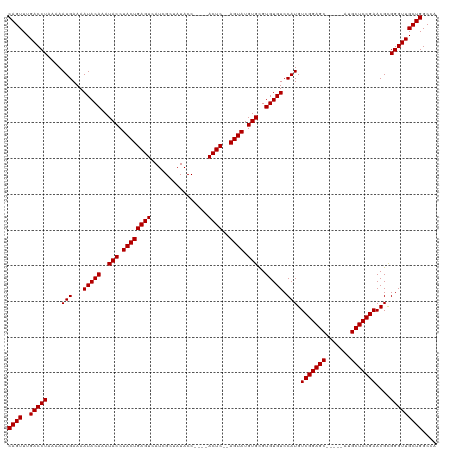
| Location | 15,876,759 – 15,876,868 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.36 |
| Shannon entropy | 0.09399 |
| G+C content | 0.47315 |
| Mean single sequence MFE | -34.12 |
| Consensus MFE | -29.30 |
| Energy contribution | -28.98 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.888824 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15876759 109 - 22422827 UGGCCAGCAACCCACGUUCGGAGCAU-----GCUCCGACAUGGACACACGCACAUACU--GAGU----GGUAUAGCGAUGACUCAGUAGGUAUUUGUAUUUUCAUUUUUAGGUCAAACUG (((((......(((.((.(((((...-----.))))))).)))(((.(..(...((((--((((----.((......)).)))))))).)..).))).............)))))..... ( -35.20, z-score = -2.89, R) >droEre2.scaffold_4690 7575303 115 + 18748788 UGGCCAGCAACCCGCGUUCGGAGCAU-----GCUCCGACAUGGACACACGCACAUACUACGAGUAUUCGGUAUAGCGAUGACUCAGUAGGUAUUUGUAUUUUCAUUUUUAGGUCAAACUG (((((......(((.((.(((((...-----.))))))).)))......(((.(((((((((((..(((......)))..)))).))).)))).))).............)))))..... ( -33.30, z-score = -2.08, R) >droYak2.chrX 10065550 120 - 21770863 UGGCCAGCAACCCGCGUUCGGAGCAUGCUCCGCUCCGACAUGGACACACGCACAUACUACGAGUAUUGGGUAUAGCGAUGACUCAGUAGGUAUUUGUAUUUUCAUUUUUAGGUCAAACUG (((((......(((.((.((((((.......)))))))).)))......(((.(((((((((((((((.......)))).)))).))).)))).))).............)))))..... ( -33.90, z-score = -1.24, R) >droSec1.super_183 1701 109 - 33538 UGGCCAGCAACCCGCGUUCGGAGCAU-----GCUCCGACAUGGACACACACACAUACU--GAGU----GGUAUAGCGAUGACUCAGUAGGUAUUUGUAUUUUCAUUUUUAGGUCAAACUG (((((......(((.((.(((((...-----.))))))).)))..(((.(.((.((((--((((----.((......)).)))))))).)).).))).............)))))..... ( -33.70, z-score = -2.42, R) >droSim1.chrX 12288367 109 - 17042790 UGGCCAGCAACCCGCGUUCGGAGCAU-----GCUCCGACAUGGACACACGCACAUACU--GAGU----GGUAUAGCGAUGACUCAGUAGGUAUUUGUAUUUUCAUUUUUAGGUCAAACUG (((((......(((.((.(((((...-----.))))))).)))(((.(..(...((((--((((----.((......)).)))))))).)..).))).............)))))..... ( -34.50, z-score = -2.32, R) >consensus UGGCCAGCAACCCGCGUUCGGAGCAU_____GCUCCGACAUGGACACACGCACAUACU__GAGU____GGUAUAGCGAUGACUCAGUAGGUAUUUGUAUUUUCAUUUUUAGGUCAAACUG (((((......(((.((.((((((.......)))))))).)))......(((.(((((..((((....(......)....))))....))))).))).............)))))..... (-29.30 = -28.98 + -0.32)

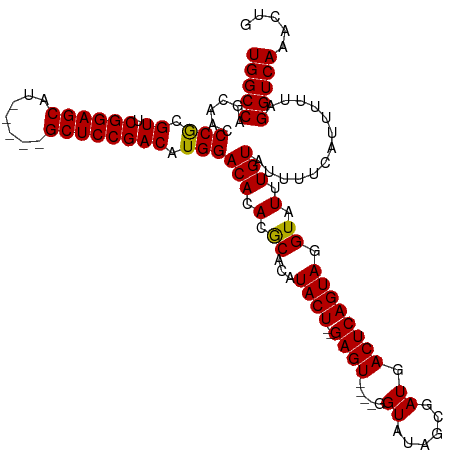
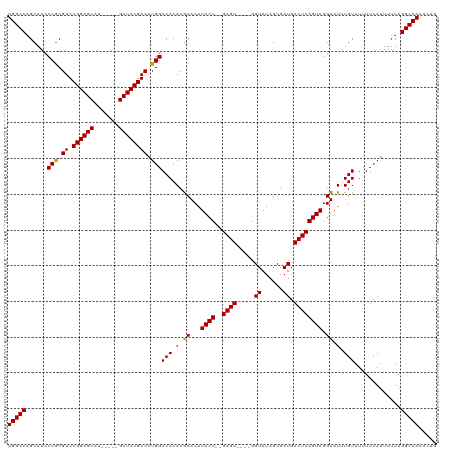
| Location | 15,876,799 – 15,876,908 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.32 |
| Shannon entropy | 0.11204 |
| G+C content | 0.52461 |
| Mean single sequence MFE | -45.90 |
| Consensus MFE | -36.88 |
| Energy contribution | -36.76 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.938903 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15876799 109 + 22422827 CAUCGCUAUACC----ACUC--AGUAUGUGCGUGUGUCCAUGUCGGA-----GCAUGCUCCGAACGUGGGUUGCUGGCCAAUUAUCUGGUUGGCAACAGGAUAUUGCAGCGAGUUUGCGU ..(((((((((.----....--.)))))((((.((((((((((((((-----(....))))).))))..(((((..((((......))))..))))).))))))))))))))........ ( -45.80, z-score = -3.05, R) >droEre2.scaffold_4690 7575343 115 - 18748788 CAUCGCUAUACCGAAUACUCGUAGUAUGUGCGUGUGUCCAUGUCGGA-----GCAUGCUCCGAACGCGGGUUGCUGGCCAAUUAUCUGGUUGGCAACAGGAUAUUGCAGCGAGUUUGCGU ..((((((((((((....)))..)))))((((.((((((...(((((-----(....))))))......(((((..((((......))))..))))).))))))))))))))........ ( -45.90, z-score = -2.55, R) >droYak2.chrX 10065590 120 + 21770863 CAUCGCUAUACCCAAUACUCGUAGUAUGUGCGUGUGUCCAUGUCGGAGCGGAGCAUGCUCCGAACGCGGGUUGCUGGCCAAUUAUCUGGUUGGCAACAGGAUAUGGCAGCGAGUUUGCGU ....((((((.((.((((.((((.....)))).))))((.((((((((((.....))))))).))).))(((((..((((......))))..))))).)).)))))).(((....))).. ( -49.60, z-score = -2.44, R) >droSec1.super_183 1741 109 + 33538 CAUCGCUAUACC----ACUC--AGUAUGUGUGUGUGUCCAUGUCGGA-----GCAUGCUCCGAACGCGGGUUGCUGGCCAAUUAUCUGGUUGGCAACAGGAUAUUGCAGCGAGUUUGUGU ..((((((((((----((..--.....))).))))((((...(((((-----(....))))))......(((((..((((......))))..))))).)))).....)))))........ ( -43.20, z-score = -2.49, R) >droSim1.chrX 12288407 109 + 17042790 CAUCGCUAUACC----ACUC--AGUAUGUGCGUGUGUCCAUGUCGGA-----GCAUGCUCCGAACGCGGGUUGCUGGCCAAUUAUCUGGUUGGCAACAGGAUAUUGCAGUGAGUUUGCGU ...(((...(((----((((--(((((.(.(((((.......(((((-----(....))))))))))).(((((..((((......))))..))))).).)))))).)))).))..))). ( -45.01, z-score = -2.82, R) >consensus CAUCGCUAUACC____ACUC__AGUAUGUGCGUGUGUCCAUGUCGGA_____GCAUGCUCCGAACGCGGGUUGCUGGCCAAUUAUCUGGUUGGCAACAGGAUAUUGCAGCGAGUUUGCGU ..(((((((((............)))))((((.((((((...(((((...........)))))......(((((..((((......))))..))))).))))))))))))))........ (-36.88 = -36.76 + -0.12)

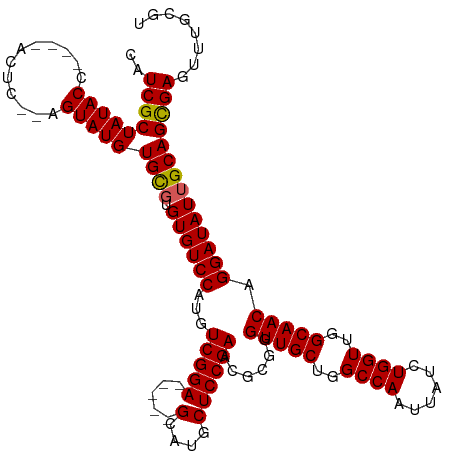

| Location | 15,876,833 – 15,876,934 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.26 |
| Shannon entropy | 0.22434 |
| G+C content | 0.59296 |
| Mean single sequence MFE | -44.50 |
| Consensus MFE | -39.82 |
| Energy contribution | -39.70 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.953091 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15876833 101 + 22422827 -----UGUCGGAGCAUGCUCCGAACGUGGGUUGCUGGCCAAUUAUCUGGUUGGCAACAGGAUAUUGCAGCGAGUUUGCGUGUCCGCGCAGCCCGACGACUCCCAGG-------------- -----..((((((....)))))).((((((((((..(((((((....)))))))....((((((.((((.....))))))))))..))))))).))).........-------------- ( -44.40, z-score = -2.57, R) >droEre2.scaffold_4690 7575383 115 - 18748788 -----UGUCGGAGCAUGCUCCGAACGCGGGUUGCUGGCCAAUUAUCUGGUUGGCAACAGGAUAUUGCAGCGAGUUUGCGUGUCCGCGCAGCACGACGACUCCCAAAUCCCAGGUCCCAAG -----..((((((....))))))....(((((((..((((......))))..))))).((((.(((..(.(((((..(((((.......)))))..)))))))))))))......))... ( -43.40, z-score = -1.46, R) >droYak2.chrX 10065630 120 + 21770863 UGUCGGAGCGGAGCAUGCUCCGAACGCGGGUUGCUGGCCAAUUAUCUGGUUGGCAACAGGAUAUGGCAGCGAGUUUGCGUGUCCGCGCAGCCCGACGACUCCCAAAUCCCAAAUCCCAGG .(((((.((((((....))))....(((.(((((..((((......))))..))))).(((((((.(((.....)))))))))).))).)))))))........................ ( -50.00, z-score = -2.15, R) >droSec1.super_183 1775 101 + 33538 -----UGUCGGAGCAUGCUCCGAACGCGGGUUGCUGGCCAAUUAUCUGGUUGGCAACAGGAUAUUGCAGCGAGUUUGUGUGUCCGCGCAGCGCGACGACUCCCAGG-------------- -----.((((..((.(((.(((....)))(((((..((((......))))..))))).((((((.((((.....))))))))))..)))..))..)))).......-------------- ( -40.60, z-score = -1.22, R) >droSim1.chrX 12288441 101 + 17042790 -----UGUCGGAGCAUGCUCCGAACGCGGGUUGCUGGCCAAUUAUCUGGUUGGCAACAGGAUAUUGCAGUGAGUUUGCGUGUCCGCGCAGCGCGACGACUCCCAGG-------------- -----.((((..((.(((.(((....)))(((((..((((......))))..))))).((((((.((((.....))))))))))..)))..))..)))).......-------------- ( -44.10, z-score = -2.26, R) >consensus _____UGUCGGAGCAUGCUCCGAACGCGGGUUGCUGGCCAAUUAUCUGGUUGGCAACAGGAUAUUGCAGCGAGUUUGCGUGUCCGCGCAGCCCGACGACUCCCAGG______________ .......((((((....))))))..(((.(((((..((((......))))..))))).((((((.((((.....)))))))))).)))................................ (-39.82 = -39.70 + -0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:48:11 2011