| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,876,370 – 15,876,449 |
| Length | 79 |
| Max. P | 0.815601 |

| Location | 15,876,370 – 15,876,449 |
|---|---|
| Length | 79 |
| Sequences | 11 |
| Columns | 86 |
| Reading direction | forward |
| Mean pairwise identity | 65.69 |
| Shannon entropy | 0.75177 |
| G+C content | 0.50558 |
| Mean single sequence MFE | -19.85 |
| Consensus MFE | -9.29 |
| Energy contribution | -8.84 |
| Covariance contribution | -0.45 |
| Combinations/Pair | 1.62 |
| Mean z-score | -0.69 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.815601 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
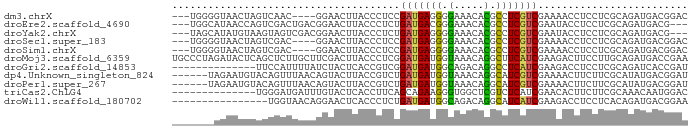
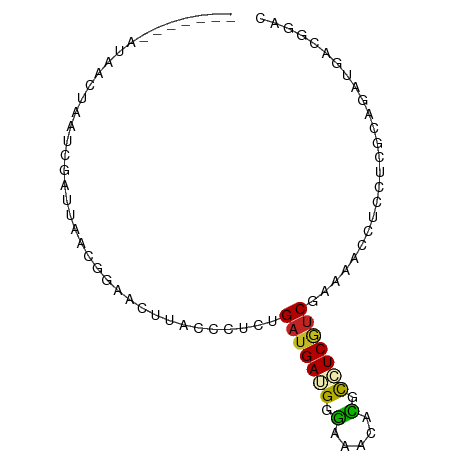
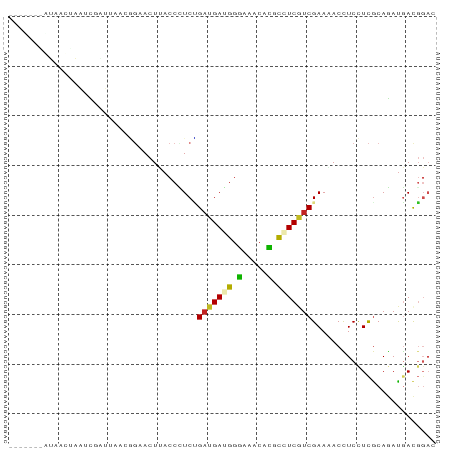
>dm3.chrX 15876370 79 + 22422827 ---UGGGGUAACUAGUCAAC----GGAACUUACCCUCCGAUGAGGGGAAACACGCCUCGUCGAAAACCUCCUCGCAGAUGACGGAC ---.(((((((....((...----.))..))))))).((((((((.(.....).))))))))......((((((....))).))). ( -24.20, z-score = -0.89, R) >droEre2.scaffold_4690 7574913 80 - 18748788 ---UGGCAUAACCAGUCGACUGACGGAACUUACCCUCUGAUGACGGGAAACACGCCUCGUCGAAUACCUCCUCGCAGAUGACG--- ---(((.....)))((((.(((.((((........)))).(((((((........)))))))............))).)))).--- ( -19.80, z-score = -1.20, R) >droYak2.chrX 10065153 80 + 21770863 ---UAGCAUAUGUAAGUAGUCGACGGAACUUACCCUCUGAUGAGGGGAAACACGCCUCGUCGAAUACCUCCUCGCAGAUGACG--- ---..((((.((..((.(((((((((.......((((....))))(....).....)))))))....)).))..)).))).).--- ( -22.00, z-score = -1.14, R) >droSec1.super_183 1311 79 + 33538 ---UGGGGUAACUAGUCGAC----GGAACUUACCCUCCGAUGAGGGGAAACACGCCUCGUCGAAAACCUCCUCGCAGAUGACGGAC ---.(((((((....((...----.))..))))))).((((((((.(.....).))))))))......((((((....))).))). ( -24.40, z-score = -0.56, R) >droSim1.chrX 12287977 79 + 17042790 ---UGGGGUAACUAGUCGAC----GGAACUUACCCUCCGAUGAGGGGAAACACGCCUCGUCGAAAACCUCCUCGCAGAUGACGGAC ---.(((((((....((...----.))..))))))).((((((((.(.....).))))))))......((((((....))).))). ( -24.40, z-score = -0.56, R) >droMoj3.scaffold_6359 2335309 86 + 4525533 UGCCCUAGAUACUCAGCUCUUGCUUCGACUUACCCUCGGAUGAUGGUAAACAGGCUUCAUCGAAGACUUCCUUGCAGAUGACCGAA .(((..........(((....))).....(((((.((....)).)))))...))).(((((.(((.....)))...)))))..... ( -15.80, z-score = 0.62, R) >droGri2.scaffold_14853 6438128 72 + 10151454 --------------UUCCAUUUUAUCUACUCACCGUCGGAUGAUGGCAGACAGGCCUCAUCGAAGACCUCCUCGCAGAUCACCGAU --------------....................(((((.((((.((((..(((.((......)).))).)).))..))))))))) ( -14.40, z-score = 0.08, R) >dp4.Unknown_singleton_824 707 80 - 5765 ------UAGAAUGUACAGUUUAACAGUACUUACCGUCUGAUGAUGGUAAACAGGCAUCGUCGAAAACUUCUUCGCAUAUGACGGAU ------......((((.(.....).))))...(((((.(((((((.(.....).)))))))(((......)))......))))).. ( -19.40, z-score = -1.54, R) >droPer1.super_267 10333 80 + 27109 ------UAGAAUGUACAGUUUAACAGUACUUACCGUCUGAUGAUGGUAAACAGGCAUCGUCGAAAACUUCUUCGCAUAUGACGGAU ------......((((.(.....).))))...(((((.(((((((.(.....).)))))))(((......)))......))))).. ( -19.40, z-score = -1.54, R) >triCas2.ChLG4 1245681 72 + 13894384 --------------UGGGAUGAUUUGUACUCACCUUCAGCAGAAGGGUGGCUCGUCUCAUCGAACACUUCUUCGCAAACAAUGGAC --------------((((((((...((...(.(((((....))))))..)))))))))).((((......))))............ ( -16.10, z-score = -0.17, R) >droWil1.scaffold_180702 3949105 70 + 4511350 ----------------UGGUAACAGGAACUCACCCUCUGAUGAUGGCAGACAGGCAUCAUCGAAGACCUCCUCACAGAUGACGGAA ----------------.(((....((......))((.((((((((.(.....).)))))))).)))))((((((....))).))). ( -18.50, z-score = -0.76, R) >consensus _______AUAACUAAUCGAUUAACGGAACUUACCCUCUGAUGAUGGGAAACACGCCUCGUCGAAAACCUCCUCGCAGAUGACGGAC ......................................(((((((.(.....).)))))))......................... ( -9.29 = -8.84 + -0.45)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:48:05 2011