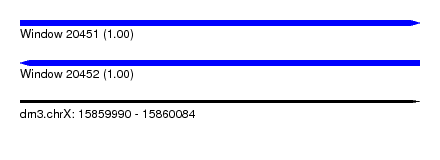
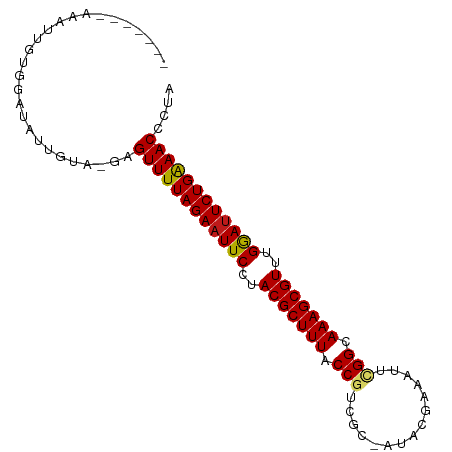
| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,859,990 – 15,860,084 |
| Length | 94 |
| Max. P | 0.999711 |

| Location | 15,859,990 – 15,860,084 |
|---|---|
| Length | 94 |
| Sequences | 14 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 76.17 |
| Shannon entropy | 0.54462 |
| G+C content | 0.39696 |
| Mean single sequence MFE | -27.58 |
| Consensus MFE | -22.24 |
| Energy contribution | -22.58 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.22 |
| Mean z-score | -4.82 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.23 |
| SVM RNA-class probability | 0.999711 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
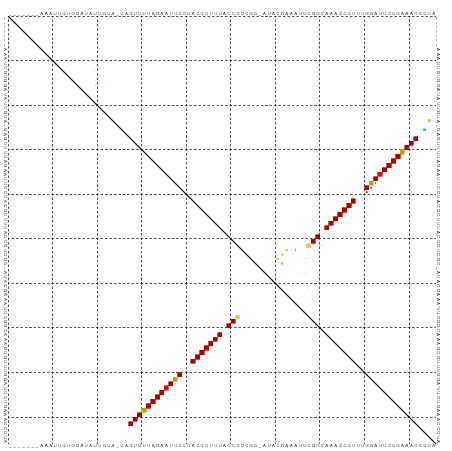
>dm3.chrX 15859990 94 + 22422827 UAGGGUUUCAGAAUCCAAACGCUUUGCCGAAUUUCGUAU-GCCACGGUAAAGCGUAGGAAUUCUAAAACUC-UACAGCAACCACAAUUUAAUAUUG ((((((((.(((((((..(((((((((((..........-....))))))))))).)).))))).))))))-))...................... ( -29.84, z-score = -5.27, R) >droSim1.chrX 12269423 94 + 17042790 UAGGGUUUCAGAAUCCAAACGCUUUGCCGAAUUUCGUAU-GCCACGGUAAAGCGUAGGAAUUCUAAAACUC-UACAGCAUCCACAAUUUAAUAUGG ((((((((.(((((((..(((((((((((..........-....))))))))))).)).))))).))))))-))...................... ( -29.84, z-score = -4.83, R) >droSec1.super_37 439560 94 + 454039 UAGGGUUUCAGAAUCCAAACGCUUUGCCGAAUUUCGUAU-GCGACGGUAAAGCGUAGGAAUUCUAAAACUC-UACAGCAUCCACAAUUUAAUAUUG ((((((((.(((((((..(((((((((((....(((...-.)))))))))))))).)).))))).))))))-))...................... ( -32.90, z-score = -5.96, R) >droEre2.scaffold_4690 7559939 94 - 18748788 UAGAGUUUCAGAAUCCAAACGCUUUGCCGAAUUUCGUAU-GCCACGGUAAAGCGUAGGAAUUCUAAAACUC-UGCAGCACCCACAAUUUAUUAUUG ((((((((.(((((((..(((((((((((..........-....))))))))))).)).))))).))))))-))...................... ( -30.44, z-score = -5.39, R) >droYak2.chrX 10049121 94 + 21770863 UAGGGUUUCAGAAUCCAAACGCUUUGCCGAAUUUCGUAU-GCGACGGUAAAGCGUAGGAAUUCUAAAACUC-UACAGCGCCCACAAUUUUUAACUG ((((((((.(((((((..(((((((((((....(((...-.)))))))))))))).)).))))).))))))-))...................... ( -32.90, z-score = -5.07, R) >droAna3.scaffold_13250 426911 88 - 3535662 UAGGGUUCCAGAAUCCAAACGCUUUGCCUAAUUUCAUAU-GCAACGGUAAAGCGUAGGAAUUCUAAAACUC-CACAAUACCCUUUUUUUA------ ..(((((..(((((((..((((((((((...........-.....)))))))))).)).)))))..))).)-).................------ ( -23.79, z-score = -4.06, R) >dp4.Unknown_group_79 8477 94 - 26009 UAGGGUUUCAGAAUCCAAACGCUUUGCCGAAUUUCGUUUUGCAACGGUAAAGCGUAGGAAUUCUAAAACAC-UACAAUAUCCUCGUUAUCUAUCU- (((.((((.(((((((..(((((((((((...............))))))))))).)).))))).)))).)-)).....................- ( -25.76, z-score = -3.89, R) >droPer1.super_192 3417 94 - 52076 UAGGGUUUCAGAAUCCAAACGCUUUGCCGAAUUUCGUUUUGCAACGGUAAAGCGUAGGAAUUCUAAAACAC-UACAAUAUCCUCGUUAUCUAUCU- (((.((((.(((((((..(((((((((((...............))))))))))).)).))))).)))).)-)).....................- ( -25.76, z-score = -3.89, R) >droWil1.scaffold_180777 2521621 86 + 4753960 UAAUGUUUCAGAAUCCAAACGCUUUGCCGAGUUUCGUAG-GCGACGGUAAAGCGUAGGAAUUCUAAAACGC-UACCAAUGUCAGAUUG-------- ....((((.(((((((..(((((((((((....(((...-.)))))))))))))).)).))))).))))..-................-------- ( -27.60, z-score = -3.58, R) >droVir3.scaffold_12970 3343541 84 + 11907090 UAUCGUUUCAGAAUCCAAACGCUUUGCCAAAUUUCGGAC-ACGACGGUAAAGCGUAGGAAUUCUAAAACGA-UACGAUGGACAUUA---------- ((((((((.(((((((..((((((((((.....(((...-.))).)))))))))).)).))))).))))))-))............---------- ( -30.30, z-score = -5.64, R) >droMoj3.scaffold_6473 7790777 81 + 16943266 UAUUGUUUCAGAAUCCAAACGCUUUGCCAAAUUGCGUAU-ACGACGGUAAAGCGUAGGAAUUCUAAAACGA-UACGCUGGCCA------------- ((((((((.(((((((..((((((((((......((...-.))..)))))))))).)).))))).))))))-)).........------------- ( -26.80, z-score = -3.84, R) >droGri2.scaffold_15203 1470469 81 - 11997470 UACCGUUUCAGAAUCCAAACGCUUUGCCAAAUUUCGUAU-ACGACGGUAAAGCGUAGGAAUUCUAAAACAA-UACAAUAGUCA------------- ....((((.(((((((..((((((((((.....(((...-.))).)))))))))).)).))))).))))..-...........------------- ( -24.60, z-score = -5.56, R) >apiMel3.Group8 3844251 81 - 11452794 ----GUUUCAGAUUUCAAACGCUUUGCC--ACUUCGAAC---AUCGGUAAAGCGUAGGAAUUCUAAAACAAAUACCACGCUUUUAUCUCG------ ----((((.(((.(((..((((((((((--.........---...))))))))))..))).))).)))).....................------ ( -23.10, z-score = -5.32, R) >anoGam1.chrX 3407734 80 - 22145176 ----GUUUCAGAAUCCAAACGCUUUGCCGCACUACUUUAAUCACGGGUAAAGCGUAGGAAUUCUAAAACAA-ACCAUUAACUAAA----------- ----((((.(((((((..((((((((((.................)))))))))).)).))))).))))..-.............----------- ( -22.43, z-score = -5.16, R) >consensus UAGGGUUUCAGAAUCCAAACGCUUUGCCGAAUUUCGUAU_GCGACGGUAAAGCGUAGGAAUUCUAAAACUC_UACAAUACCCACAAUUU_______ ..((((((.(((((((..((((((((((.................)))))))))).)).))))).))))))......................... (-22.24 = -22.58 + 0.34)

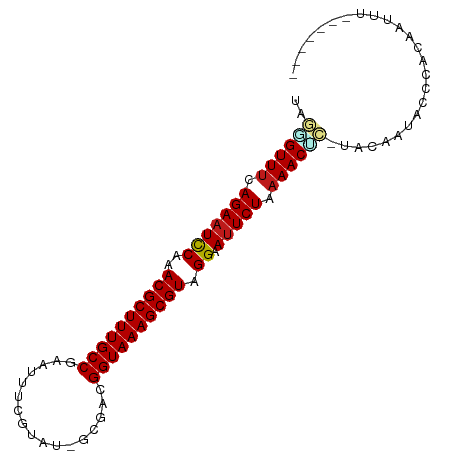
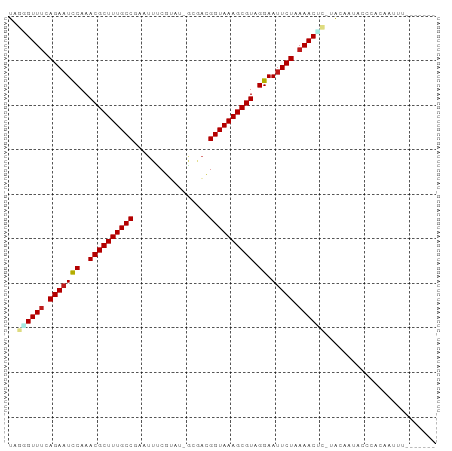
| Location | 15,859,990 – 15,860,084 |
|---|---|
| Length | 94 |
| Sequences | 14 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 76.17 |
| Shannon entropy | 0.54462 |
| G+C content | 0.39696 |
| Mean single sequence MFE | -28.88 |
| Consensus MFE | -25.28 |
| Energy contribution | -25.31 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.14 |
| Mean z-score | -4.08 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.90 |
| SVM RNA-class probability | 0.999450 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15859990 94 - 22422827 CAAUAUUAAAUUGUGGUUGCUGUA-GAGUUUUAGAAUUCCUACGCUUUACCGUGGC-AUACGAAAUUCGGCAAAGCGUUUGGAUUCUGAAACCCUA ......................((-(.((((((((((((..(((((((.(((....-..........))).)))))))..)))))))))))).))) ( -28.34, z-score = -3.27, R) >droSim1.chrX 12269423 94 - 17042790 CCAUAUUAAAUUGUGGAUGCUGUA-GAGUUUUAGAAUUCCUACGCUUUACCGUGGC-AUACGAAAUUCGGCAAAGCGUUUGGAUUCUGAAACCCUA (((((......)))))......((-(.((((((((((((..(((((((.(((....-..........))).)))))))..)))))))))))).))) ( -32.94, z-score = -4.87, R) >droSec1.super_37 439560 94 - 454039 CAAUAUUAAAUUGUGGAUGCUGUA-GAGUUUUAGAAUUCCUACGCUUUACCGUCGC-AUACGAAAUUCGGCAAAGCGUUUGGAUUCUGAAACCCUA ......................((-(.((((((((((((..(((((((.((((((.-...)))....))).)))))))..)))))))))))).))) ( -29.90, z-score = -4.15, R) >droEre2.scaffold_4690 7559939 94 + 18748788 CAAUAAUAAAUUGUGGGUGCUGCA-GAGUUUUAGAAUUCCUACGCUUUACCGUGGC-AUACGAAAUUCGGCAAAGCGUUUGGAUUCUGAAACUCUA ............(..(...)..)(-((((((((((((((..(((((((.(((....-..........))).)))))))..))))))))))))))). ( -33.04, z-score = -4.51, R) >droYak2.chrX 10049121 94 - 21770863 CAGUUAAAAAUUGUGGGCGCUGUA-GAGUUUUAGAAUUCCUACGCUUUACCGUCGC-AUACGAAAUUCGGCAAAGCGUUUGGAUUCUGAAACCCUA ((((..............))))((-(.((((((((((((..(((((((.((((((.-...)))....))).)))))))..)))))))))))).))) ( -30.44, z-score = -3.25, R) >droAna3.scaffold_13250 426911 88 + 3535662 ------UAAAAAAAGGGUAUUGUG-GAGUUUUAGAAUUCCUACGCUUUACCGUUGC-AUAUGAAAUUAGGCAAAGCGUUUGGAUUCUGGAACCCUA ------.................(-(.((((((((((((..(((((((.((.....-...........)).)))))))..)))))))))))))).. ( -27.29, z-score = -3.31, R) >dp4.Unknown_group_79 8477 94 + 26009 -AGAUAGAUAACGAGGAUAUUGUA-GUGUUUUAGAAUUCCUACGCUUUACCGUUGCAAAACGAAAUUCGGCAAAGCGUUUGGAUUCUGAAACCCUA -.........((((.....))))(-(.((((((((((((..(((((((.(((...............))).)))))))..)))))))))))).)). ( -28.36, z-score = -3.75, R) >droPer1.super_192 3417 94 + 52076 -AGAUAGAUAACGAGGAUAUUGUA-GUGUUUUAGAAUUCCUACGCUUUACCGUUGCAAAACGAAAUUCGGCAAAGCGUUUGGAUUCUGAAACCCUA -.........((((.....))))(-(.((((((((((((..(((((((.(((...............))).)))))))..)))))))))))).)). ( -28.36, z-score = -3.75, R) >droWil1.scaffold_180777 2521621 86 - 4753960 --------CAAUCUGACAUUGGUA-GCGUUUUAGAAUUCCUACGCUUUACCGUCGC-CUACGAAACUCGGCAAAGCGUUUGGAUUCUGAAACAUUA --------................-..((((((((((((..(((((((.((((((.-...)))....))).)))))))..)))))))))))).... ( -29.10, z-score = -4.52, R) >droVir3.scaffold_12970 3343541 84 - 11907090 ----------UAAUGUCCAUCGUA-UCGUUUUAGAAUUCCUACGCUUUACCGUCGU-GUCCGAAAUUUGGCAAAGCGUUUGGAUUCUGAAACGAUA ----------............((-((((((((((((((..(((((((.((((((.-...)))....))).)))))))..)))))))))))))))) ( -32.40, z-score = -6.00, R) >droMoj3.scaffold_6473 7790777 81 - 16943266 -------------UGGCCAGCGUA-UCGUUUUAGAAUUCCUACGCUUUACCGUCGU-AUACGCAAUUUGGCAAAGCGUUUGGAUUCUGAAACAAUA -------------...........-..((((((((((((..(((((((.(((.((.-...)).....))).)))))))..)))))))))))).... ( -26.60, z-score = -3.58, R) >droGri2.scaffold_15203 1470469 81 + 11997470 -------------UGACUAUUGUA-UUGUUUUAGAAUUCCUACGCUUUACCGUCGU-AUACGAAAUUUGGCAAAGCGUUUGGAUUCUGAAACGGUA -------------.........((-((((((((((((((..(((((((.((((((.-...)))....))).)))))))..)))))))))))))))) ( -30.70, z-score = -5.93, R) >apiMel3.Group8 3844251 81 + 11452794 ------CGAGAUAAAAGCGUGGUAUUUGUUUUAGAAUUCCUACGCUUUACCGAU---GUUCGAAGU--GGCAAAGCGUUUGAAAUCUGAAAC---- ------.....................((((((((.(((..(((((((.(((.(---......).)--)).)))))))..))).))))))))---- ( -21.00, z-score = -2.11, R) >anoGam1.chrX 3407734 80 + 22145176 -----------UUUAGUUAAUGGU-UUGUUUUAGAAUUCCUACGCUUUACCCGUGAUUAAAGUAGUGCGGCAAAGCGUUUGGAUUCUGAAAC---- -----------.............-..((((((((((((..(((((((..((((.((((...)))))))).)))))))..))))))))))))---- ( -25.80, z-score = -4.07, R) >consensus _______AAAUUGUGGAUAUUGUA_GAGUUUUAGAAUUCCUACGCUUUACCGUCGC_AUACGAAAUUCGGCAAAGCGUUUGGAUUCUGAAACCCUA ...........................((((((((((((..(((((((.(((...............))).)))))))..)))))))))))).... (-25.28 = -25.31 + 0.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:48:05 2011