| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,480,282 – 11,480,372 |
| Length | 90 |
| Max. P | 0.753158 |

| Location | 11,480,282 – 11,480,372 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 61.58 |
| Shannon entropy | 0.68902 |
| G+C content | 0.32013 |
| Mean single sequence MFE | -14.54 |
| Consensus MFE | -6.22 |
| Energy contribution | -7.26 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.742344 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
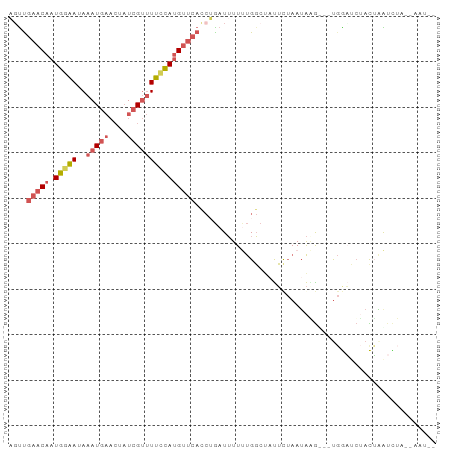
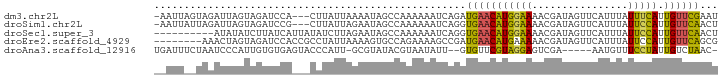
>dm3.chr2L 11480282 90 + 23011544 AUUCGAACAAUGAAAUAAAUGAACUAUCGUUUUCCAUGUUCAUCUGAUUUUUUGGCUAUUUUAAUAAG---UGGAUCUACUAAUCUACUAAUU- ....(((((.((....((((((....))))))..))))))).........................((---(((((......)))))))....- ( -13.90, z-score = -1.49, R) >droSim1.chr2L 11293463 90 + 22036055 AGUUGAACAAUGGAAUAAAUGAACUAUCGUUUUCCAUGUUCACCUGAUUUUUUGGCUAUUCUAAUAAG---CGGAUCUACUAAUCUAAUAAUU- ((.((((((.(((((..(((((....)))))))))))))))).))((((..((.(((.........))---).))......))))........- ( -17.10, z-score = -1.91, R) >droSec1.super_3 6875285 84 + 7220098 AGUUGAACAAUGGAAUAAAUGAACUAUCGUUUUCCAUGUUCACCUGAUUUUUUGGCUAUUCUAAGAUAUAAUGAUAAGAUAUAU---------- ((.((((((.(((((..(((((....)))))))))))))))).))....((((((.....))))))..................---------- ( -15.00, z-score = -1.19, R) >droEre2.scaffold_4929 12688256 86 - 26641161 CGCUGAACAAUGGAAUAAAUGAACUAUCGUUUUUCAUGUUCAUCGGCUUUUCUGGCACUUUUAAUAGGCGGUGGAUCUACUAGUUU-------- (((((((((.(((((..(((((....)))))))))))))))....(((.....)))..........))))................-------- ( -16.90, z-score = -0.79, R) >droAna3.scaffold_12916 12182921 85 - 16180835 -GUUAGACAAUAGGAAACAUU-----UCGACUCCUACGAACAC--AAUAUUACGUAUACGC-AAUGGGUACUCACACAAUGGGAUUAGAAAUCA -...........(....)(((-----((...(((((.......--..((((.((....)).-)))).((......))..)))))...))))).. ( -9.80, z-score = 0.34, R) >consensus AGUUGAACAAUGGAAUAAAUGAACUAUCGUUUUCCAUGUUCACCUGAUUUUUUGGCUAUUCUAAUAAG___UGGAUCUACUAAUCUA__AAU__ ....(((((.(((((..(((((....)))))))))))))))..................................................... ( -6.22 = -7.26 + 1.04)


| Location | 11,480,282 – 11,480,372 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 61.58 |
| Shannon entropy | 0.68902 |
| G+C content | 0.32013 |
| Mean single sequence MFE | -11.58 |
| Consensus MFE | -6.89 |
| Energy contribution | -6.25 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.55 |
| Mean z-score | -0.31 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.753158 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
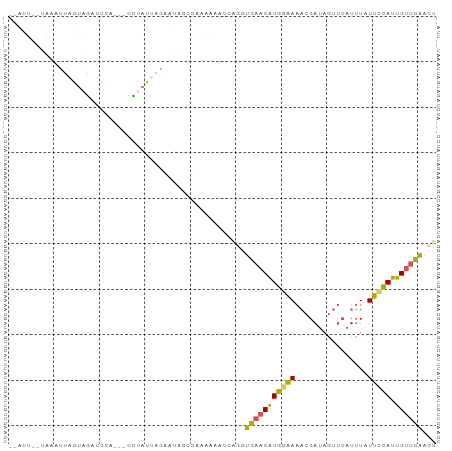
>dm3.chr2L 11480282 90 - 23011544 -AAUUAGUAGAUUAGUAGAUCCA---CUUAUUAAAAUAGCCAAAAAAUCAGAUGAACAUGGAAAACGAUAGUUCAUUUAUUUCAUUGUUCGAAU -(((.(((.((((....)))).)---)).)))........(((..((..((((((((((.(....).)).))))))))..))..)))....... ( -10.60, z-score = -0.09, R) >droSim1.chr2L 11293463 90 - 22036055 -AAUUAUUAGAUUAGUAGAUCCG---CUUAUUAGAAUAGCCAAAAAAUCAGGUGAACAUGGAAAACGAUAGUUCAUUUAUUCCAUUGUUCAACU -..........(((((((.....---.)))))))....(((.........)))(((((((((((((....)))......))))).))))).... ( -12.50, z-score = -0.36, R) >droSec1.super_3 6875285 84 - 7220098 ----------AUAUAUCUUAUCAUUAUAUCUUAGAAUAGCCAAAAAAUCAGGUGAACAUGGAAAACGAUAGUUCAUUUAUUCCAUUGUUCAACU ----------............................(((.........)))(((((((((((((....)))......))))).))))).... ( -11.00, z-score = -0.59, R) >droEre2.scaffold_4929 12688256 86 + 26641161 --------AAACUAGUAGAUCCACCGCCUAUUAAAAGUGCCAGAAAAGCCGAUGAACAUGAAAAACGAUAGUUCAUUUAUUCCAUUGUUCAGCG --------....((((((.........))))))..............((...((((((.((((((.((....)).))).)))...)))))))). ( -9.80, z-score = -0.15, R) >droAna3.scaffold_12916 12182921 85 + 16180835 UGAUUUCUAAUCCCAUUGUGUGAGUACCCAUU-GCGUAUACGUAAUAUU--GUGUUCGUAGGAGUCGA-----AAUGUUUCCUAUUGUCUAAC- .((((((((....(((.((((..(((..(...-..)..)))...)))).--)))....))))))))..-----....................- ( -14.00, z-score = -0.36, R) >consensus __AUU__UAAAUUAGUAGAUCCA___CCUAUUAGAAUAGCCAAAAAAUCAGGUGAACAUGGAAAACGAUAGUUCAUUUAUUCCAUUGUUCAACU ....................................................(((((((((((................))))).))))))... ( -6.89 = -6.25 + -0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:32:47 2011