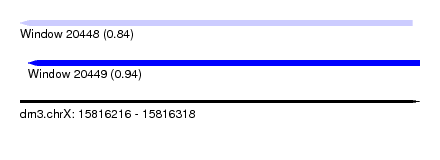
| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,816,216 – 15,816,318 |
| Length | 102 |
| Max. P | 0.943772 |

| Location | 15,816,216 – 15,816,316 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 82.88 |
| Shannon entropy | 0.30553 |
| G+C content | 0.34248 |
| Mean single sequence MFE | -20.97 |
| Consensus MFE | -16.20 |
| Energy contribution | -16.76 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.844873 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
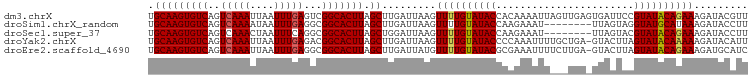
>dm3.chrX 15816216 100 - 22422827 CAAGUGUCAGUCAAAUUAAUUUGAGUCGGCACUUAGCUUGAUUAAGUUUUGUAUACCACAAAAUUAGUUGAGUGAUUCCGUAUACAGAAAGAUACGUUCA .((((((((.(((((....))))).).))))))).(((..(((((.((((((.....)))))))))))..)))((...(((((........))))).)). ( -26.10, z-score = -3.41, R) >droSim1.chrX_random 4083972 92 - 5698898 CAAGUGUCAGUCAAAAUAAUUUGAGGCGGCACUUAGCUUGAUUAAGUUUUGUAUACCAAGAAAU--------UUAGUAGGUAUGCAUAAAGAUACCUUCA .(((((((.(((..((....))..))))))))))......((((((((((.........)))))--------)))))((((((........))))))... ( -19.00, z-score = -1.26, R) >droSec1.super_37 392497 92 - 454039 CAAGUGUCAGUCAAACUAAUUUCAGGCGGCACUUAGCUGGAUUAAGUUUUGUAUACCAAGAAAU--------UUAGUACGUAUACAGAAAGAUACCUUCA .(((((((.(((............)))))))))).(..(((((...((((((((((........--------.......)))))))))).))).))..). ( -19.06, z-score = -1.48, R) >droYak2.chrX 10006894 99 - 21770863 CAAGUGUCAGUCAAAUUAAUUUGAGACGGCACUUAGCUUGAUUAAGUUUUGUAUACCCCAAAUUUUGCUGA-GUACUUAGUAUACAAAAAGAUACAUUCA ...(((((.....((((((.(((((......))))).))))))...((((((((((.....((((....))-)).....)))))))))).)))))..... ( -19.60, z-score = -1.48, R) >droEre2.scaffold_4690 7516820 99 + 18748788 CAAGUGUCAGUCAAAUUAAUUUGAGGCGGCACUUAGCUUGAUUAUGUUUUGUAUACGCGAAAUUUUCUUGA-GUACUUAGUAUACAGAAAGAUGCAUCCA ...(((((.....((((((.(((((......))))).))))))...((((((((((.....((((....))-)).....)))))))))).)))))..... ( -21.10, z-score = -0.75, R) >consensus CAAGUGUCAGUCAAAUUAAUUUGAGGCGGCACUUAGCUUGAUUAAGUUUUGUAUACCAAAAAAUU___UGA_UUACUAAGUAUACAGAAAGAUACAUUCA ...(((((.....((((((.(((((......))))).))))))...((((((((((.......................)))))))))).)))))..... (-16.20 = -16.76 + 0.56)

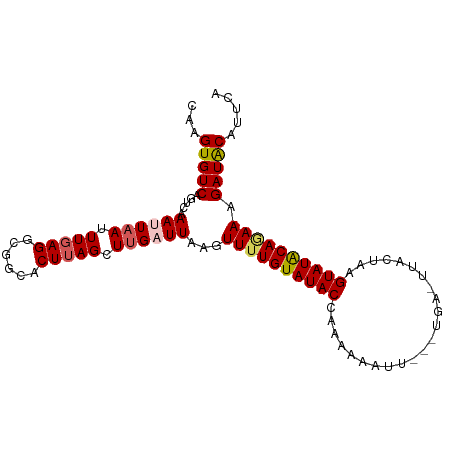
| Location | 15,816,218 – 15,816,318 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 82.88 |
| Shannon entropy | 0.30553 |
| G+C content | 0.34248 |
| Mean single sequence MFE | -22.27 |
| Consensus MFE | -18.42 |
| Energy contribution | -18.54 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.943772 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15816218 100 - 22422827 UGCAAGUGUCAGUCAAAUUAAUUUGAGUCGGCACUUAGCUUGAUUAAGUUUUGUAUACCACAAAAUUAGUUGAGUGAUUCCGUAUACAGAAAGAUACGUU ...((((((((.(((((....))))).).))))))).(((..(((((.((((((.....)))))))))))..))).....(((((........))))).. ( -25.60, z-score = -2.99, R) >droSim1.chrX_random 4083974 92 - 5698898 UGCAAGUGUCAGUCAAAAUAAUUUGAGGCGGCACUUAGCUUGAUUAAGUUUUGUAUACCAAGAAAU--------UUAGUAGGUAUGCAUAAAGAUACCUU .(((((((((.(((..((....))..)))))))))).))...((((((((((.........)))))--------)))))((((((........)))))). ( -21.40, z-score = -1.80, R) >droSec1.super_37 392499 92 - 454039 UGCAAGUGUCAGUCAAACUAAUUUCAGGCGGCACUUAGCUGGAUUAAGUUUUGUAUACCAAGAAAU--------UUAGUACGUAUACAGAAAGAUACCUU .(((((((((.(((............)))))))))).)).(((((...((((((((((........--------.......)))))))))).))).)).. ( -21.06, z-score = -1.82, R) >droYak2.chrX 10006896 99 - 21770863 UGCAAGUGUCAGUCAAAUUAAUUUGAGACGGCACUUAGCUUGAUUAAGUUUUGUAUACCCCAAAUUUUGCUGA-GUACUUAGUAUACAAAAAGAUACAUU .(((((((((.(((............)))))))))).)).........((((((((((.....((((....))-)).....))))))))))......... ( -20.90, z-score = -1.69, R) >droEre2.scaffold_4690 7516822 99 + 18748788 UGCAAGUGUCAGUCAAAUUAAUUUGAGGCGGCACUUAGCUUGAUUAUGUUUUGUAUACGCGAAAUUUUCUUGA-GUACUUAGUAUACAGAAAGAUGCAUC .(((((((((..(((((....)))))...))))))).)).((......((((((((((.....((((....))-)).....)))))))))).....)).. ( -22.40, z-score = -0.85, R) >consensus UGCAAGUGUCAGUCAAAUUAAUUUGAGGCGGCACUUAGCUUGAUUAAGUUUUGUAUACCAAAAAAUU___UGA_UUACUAAGUAUACAGAAAGAUACAUU .(((((((((..(((((....)))))...))))))).)).........((((((((((.......................))))))))))......... (-18.42 = -18.54 + 0.12)
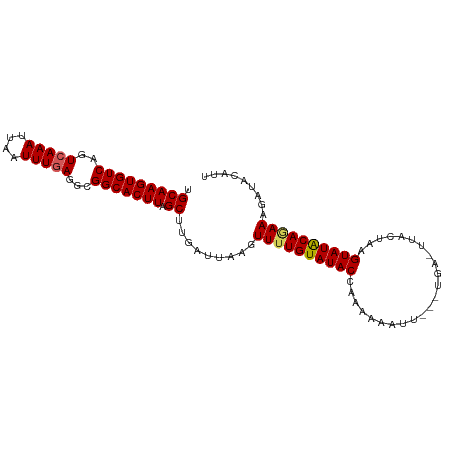
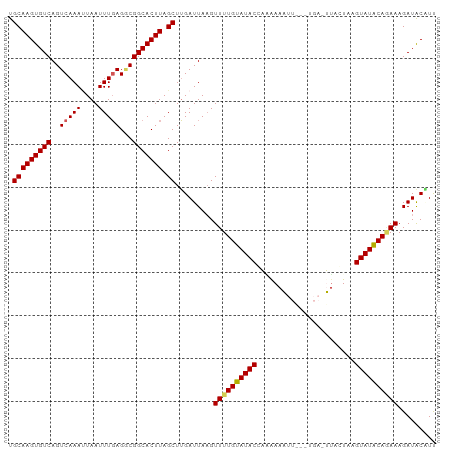
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:48:02 2011