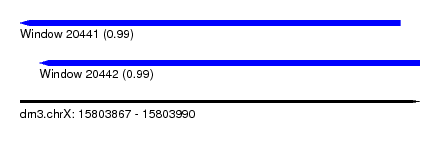
| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,803,867 – 15,803,990 |
| Length | 123 |
| Max. P | 0.993142 |

| Location | 15,803,867 – 15,803,984 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 78.30 |
| Shannon entropy | 0.35232 |
| G+C content | 0.31483 |
| Mean single sequence MFE | -26.10 |
| Consensus MFE | -15.30 |
| Energy contribution | -15.93 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.36 |
| SVM RNA-class probability | 0.989263 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
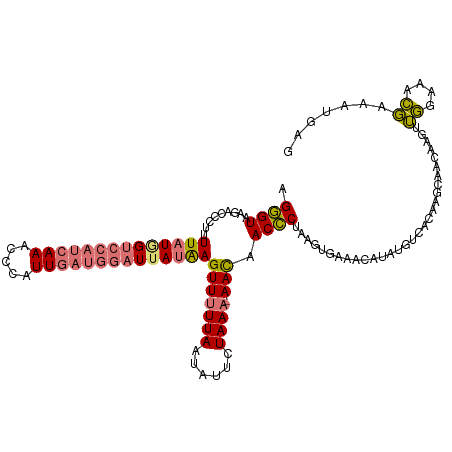
>dm3.chrX 15803867 117 - 22422827 AGGGUAAGACCCUUUUAUAGUCCAUCAAACCCAUUGAUGGAUUAUAAGUUUUUAAUGUUCUAAU-AACAACCCUAAGUGAAACAUAUUUCACAAGCAACAAGUUGGAAACGCAAUGAG ((((.....)))).((((((((((((((.....))))))))))))))((((((((((((.....-)))........((((((....)))))).........)))))))))........ ( -31.70, z-score = -4.36, R) >droSim1.chrX 12222964 117 - 17042790 CGGGUAAGACCGUUUUAUGGUCCAUCAAAUCCAUUGAUGGAUUAUAAGUUUUUAAUAUUCUAAA-AACAACCCUAAUUAAAACAUAUGUUACAAGCAACAAGUUGGAAUCGAAAUGAG .((((.........((((((((((((((.....))))))))))))))(((((((......))))-))).)))).........((..((((......))))...))............. ( -27.00, z-score = -2.83, R) >droSec1.super_37 380292 117 - 454039 AGGGUAAGACAGUUUUAUGGUCCAUCAAGCCCAUUGAUGGAUUAUAAGUUUUUAAUAUUCUAAA-AACAACCCUAAGUGAAACAUAUGUUACAAGCAACAAGUUGGAAACGAAAUGAG (((((.........((((((((((((((.....))))))))))))))(((((((......))))-))).)))))..((((........))))......((..(((....)))..)).. ( -33.40, z-score = -4.62, R) >droYak2.chrX 9994789 112 - 21770863 AGUGU-UUCCUCUUUUAUUGUGCAUCAAACCCAUU-----UUUAAGAGUUUUUAUUAUUCUAAAGAAUAACACUCUAUGCUAGACAUUUCACAAAAAAACAGCAUGAAAAUAAAUAAG (((((-...(((((.....(((.........))).-----...))))).......(((((....)))))))))).((((((...................))))))............ ( -12.31, z-score = 0.08, R) >consensus AGGGUAAGACCCUUUUAUGGUCCAUCAAACCCAUUGAUGGAUUAUAAGUUUUUAAUAUUCUAAA_AACAACCCUAAGUGAAACAUAUGUCACAAGCAACAAGUUGGAAACGAAAUGAG .((((.........((((((((((((((.....))))))))))))))(((((((......)))).))).))))..............................((....))....... (-15.30 = -15.93 + 0.62)

| Location | 15,803,873 – 15,803,990 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 77.30 |
| Shannon entropy | 0.36767 |
| G+C content | 0.30414 |
| Mean single sequence MFE | -26.05 |
| Consensus MFE | -15.30 |
| Energy contribution | -15.93 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.22 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.59 |
| SVM RNA-class probability | 0.993142 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
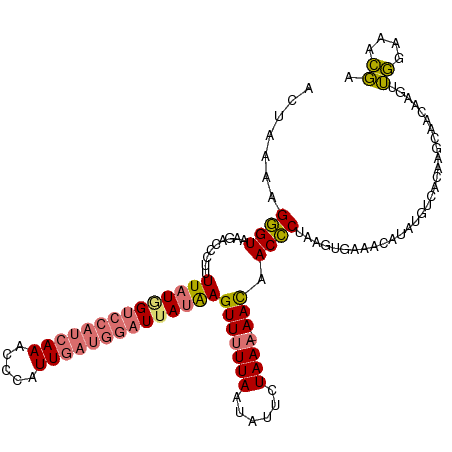
>dm3.chrX 15803873 117 - 22422827 AUUAUAAGGGUAAGACCCUUUUAUAGUCCAUCAAACCCAUUGAUGGAUUAUAAGUUUUUAAUGUUCUAAU-AACAACCCUAAGUGAAACAUAUUUCACAAGCAACAAGUUGGAAACGC .....(((((.....)))))((((((((((((((.....))))))))))))))((((((((((((.....-)))........((((((....)))))).........))))))))).. ( -32.80, z-score = -5.03, R) >droSim1.chrX 12222970 117 - 17042790 AUUAAACGGGUAAGACCGUUUUAUGGUCCAUCAAAUCCAUUGAUGGAUUAUAAGUUUUUAAUAUUCUAAA-AACAACCCUAAUUAAAACAUAUGUUACAAGCAACAAGUUGGAAUCGA ....(((((((.........((((((((((((((.....))))))))))))))(((((((......))))-))).)))).............((((......)))).)))........ ( -27.60, z-score = -3.11, R) >droSec1.super_37 380298 117 - 454039 ACUAAAAGGGUAAGACAGUUUUAUGGUCCAUCAAGCCCAUUGAUGGAUUAUAAGUUUUUAAUAUUCUAAA-AACAACCCUAAGUGAAACAUAUGUUACAAGCAACAAGUUGGAAACGA ......(((((.........((((((((((((((.....))))))))))))))(((((((......))))-))).)))))..((((........))))..........(((....))) ( -31.20, z-score = -3.82, R) >droYak2.chrX 9994795 112 - 21770863 ACAAAAAGUGU-UUCCUCUUUUAUUGUGCAUCAAACCCAUU-----UUUAAGAGUUUUUAUUAUUCUAAAGAAUAACACUCUAUGCUAGACAUUUCACAAAAAAACAGCAUGAAAAUA ......(((((-...(((((.....(((.........))).-----...))))).......(((((....)))))))))).((((((...................))))))...... ( -12.61, z-score = -0.04, R) >consensus ACUAAAAGGGUAAGACCCUUUUAUGGUCCAUCAAACCCAUUGAUGGAUUAUAAGUUUUUAAUAUUCUAAA_AACAACCCUAAGUGAAACAUAUGUCACAAGCAACAAGUUGGAAACGA .......((((.........((((((((((((((.....))))))))))))))(((((((......)))).))).))))..............................((....)). (-15.30 = -15.93 + 0.62)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:47:56 2011