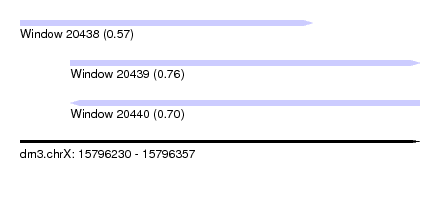
| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,796,230 – 15,796,357 |
| Length | 127 |
| Max. P | 0.763739 |

| Location | 15,796,230 – 15,796,323 |
|---|---|
| Length | 93 |
| Sequences | 13 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 73.94 |
| Shannon entropy | 0.53976 |
| G+C content | 0.66333 |
| Mean single sequence MFE | -37.15 |
| Consensus MFE | -18.32 |
| Energy contribution | -18.23 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.568173 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
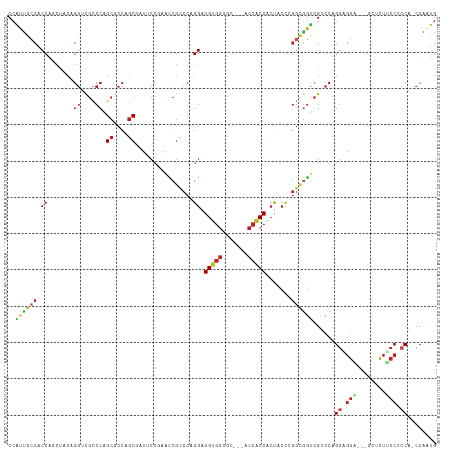
>dm3.chrX 15796230 93 + 22422827 ---------------GGGUGGACGGAACCCACCAUUGCCACCAUCUACAGGUCGCCCAGCGCCAGCGAUUCCGAAUCGCCCAGGAGGUGGGGC---ACCACCACUACCCGG ---------------((((((..((......))..((((.((((((.(.((.(((...))))).(((((.....)))))...).)))))))))---)......)))))).. ( -36.60, z-score = -1.39, R) >droSim1.chrX 12215126 93 + 17042790 ---------------GGGUGGACGGAACCCACCAUUGCCACCAUCUACAGGUCGCCCAGCGCCAGCGAUUCCGAAUCGCCCAGGAGGUGGGGC---ACCACCACUACCCGG ---------------((((((..((......))..((((.((((((.(.((.(((...))))).(((((.....)))))...).)))))))))---)......)))))).. ( -36.60, z-score = -1.39, R) >droSec1.super_37 372467 93 + 454039 ---------------GGGUGGACGGAACCCACCAUUGCCACCAUCUACAGGUCGCCCAGCGCCAGCGAUUCCGAAUCGCCCAGGAGGUGGCGC---ACCACCACUACCCGG ---------------((((((..((......))...((((((.(((...((.(((...))))).(((((.....)))))...)))))))))..---.......)))))).. ( -36.20, z-score = -2.01, R) >droYak2.chrX 9987273 93 + 21770863 ---------------GGGUGGACGAAACCCACCAUUGCCACCAGCUACAGGUCGCCCAGCGCCAGCGAUUCCGAAUCGCCCAGGAGGUGGGGC---ACCACCACUACCCGG ---------------((((((......((((((....((..........((.(((...))))).(((((.....)))))...)).))))))..---.......)))))).. ( -32.76, z-score = -0.57, R) >droEre2.scaffold_4690 7497350 93 - 18748788 ---------------GGGUGGACGGAACCCACCAUUGCCACCAGCAACAGGGCGCCCAGCGCCAGCGAUUCCGAAUCGCCCAGGAGGUGGGGC---ACCACCACUACCCGG ---------------((((((..((..((((((.((((.....))))...(((((...))))).(((((.....)))))......))))))..---.))....)))))).. ( -39.40, z-score = -1.79, R) >droAna3.scaffold_13417 5145288 93 + 6960332 ---------------AGGAGGACGAAACCCCCCGUUGCCUCCAUCUACAGGUCGCCCAGCUCCAGCGAUUCCGAAUCGUCCAGGAGGUGGAGC---CCCACCUCUUCCUGG ---------------((((((((((........((.((((........)))).))...((....)).........)))))).((((((((...---.)))))))))))).. ( -33.50, z-score = -1.61, R) >dp4.Unknown_group_88 14493 93 - 30850 ---------------GGGUGGGCGAAAUCCCCCAUUACCACCGGCAACUGGGCGCCCAGCGCCAGCUAUUCCCAAUCGCCCAGGAGGUGGUGC---ACCACCACUACCUGG ---------------(((((((.(....).))))........(((..((((....)))).))).......)))......(((((.(((((((.---..))))))).))))) ( -38.90, z-score = -1.44, R) >droPer1.super_141 49289 93 + 87512 ---------------GGGUGGGCGAAAUCCCCCAUUACCACCGGCAACUGGGCGCCCAGCGCCAGCUAUUCCCAAUCGCCCAGGAGGUGGUGC---ACCACCACUACCUGG ---------------(((((((.(....).))))........(((..((((....)))).))).......)))......(((((.(((((((.---..))))))).))))) ( -38.90, z-score = -1.44, R) >droWil1.scaffold_181096 6530755 93 - 12416693 ---------------GGGUGGACGAAAUCCACCAUUGCCACCGGCAACUGGUCGCCCAGCACCAGCAAUUCCAAGUCGUCCUGGAGGCGGAGC---ACCUCCACUACCUGG ---------------((((((((((.........(((((...)))))(((((.((...)))))))..........))))))((((((......---.))))))..)))).. ( -32.80, z-score = -1.04, R) >droMoj3.scaffold_6359 2223410 93 + 4525533 ---------------GGGUGGCCGUAAUCCACCUUUGCCACCGGCAACGGGUCGUCCAGCGCCCGCUAUUCCCAGCCGCCCAGGAGGCGGUGC---UCCGCCCCUGCCUGG ---------------(((((((............(((((...)))))((((.((.....)))))).........)))))))(((.(((((...---.))))))))...... ( -41.70, z-score = -1.22, R) >droGri2.scaffold_15203 5321936 93 - 11997470 ---------------GGGUGGACGCAAUCCACCAUUGCCACCGGCAACUGGACGCCCAGCGCCAGCAAUACCCAGCCGCCCAGGAGGUGGUGC---ACCACCGCUACCAGG ---------------((((((.((((((.....)))))....(((..((((....)))).)))...........))))))).((.(((((((.---..))))))).))... ( -37.30, z-score = -1.33, R) >anoGam1.chr2R 31534269 111 - 62725911 CUCUAUGGGAGCUCCGGCUGGCCGGGGACCACCACCAGCACCAGCUAGUGCUAGGCCGGCUCCGGCAAUACCGAACCGACCUGGAGGUGGUGCCGCACCACCCCUGCCCGG .....((((.(((..((((((((((((....)).))(((((......))))).))))))))..)))................((.(((((((...)))))))))..)))). ( -49.20, z-score = -0.63, R) >apiMel3.Group3 10461128 102 - 12341916 ------UGGUGGACCAAGACGUGGAGUGACACAGCCACCCCCUCUUUCUAGUUCUCGAGCUCCACC---UCCAGUUCCUACGGGUGGCCGACCAGCGCCGGCUAUUCCUAA ------.((((((..((((.(.((.(((.......)))))).))))....(((....)))))))))---............(((((((((........))))))))).... ( -29.10, z-score = 0.49, R) >consensus _______________GGGUGGACGAAACCCACCAUUGCCACCAGCUACAGGUCGCCCAGCGCCAGCGAUUCCGAAUCGCCCAGGAGGUGGUGC___ACCACCACUACCCGG ...............((((((...................((.......((....)).((....))................)).(((((.......))))).)))))).. (-18.32 = -18.23 + -0.09)

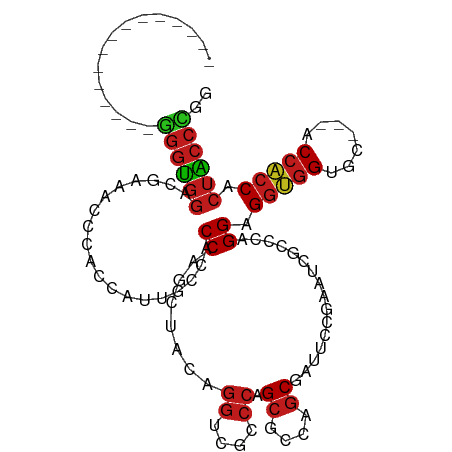

| Location | 15,796,246 – 15,796,357 |
|---|---|
| Length | 111 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 75.70 |
| Shannon entropy | 0.53857 |
| G+C content | 0.66231 |
| Mean single sequence MFE | -42.76 |
| Consensus MFE | -19.04 |
| Energy contribution | -19.48 |
| Covariance contribution | 0.43 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.763739 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
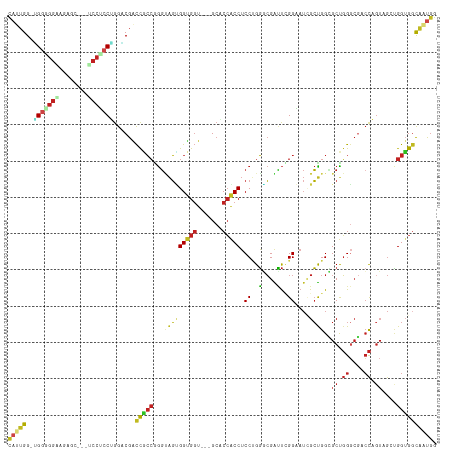
>dm3.chrX 15796246 111 + 22422827 CCAUUGCCACCAUCUACAGGUCGCCCAGCGCCAGCGAUUCCGAAUCGCCCAGGAGGUGGGGC---ACCACCACUACCCGGCGGUCGUCCCGGAGGA---UCUCUUCCCCCA-CCAAUG .(((((..(((.(((...((.(((...))))).(((((.....)))))...))))))(((((---(((.((.......)).))).)))))((.(((---.....))).)).-.))))) ( -38.40, z-score = -0.98, R) >droSim1.chrX 12215142 111 + 17042790 CCAUUGCCACCAUCUACAGGUCGCCCAGCGCCAGCGAUUCCGAAUCGCCCAGGAGGUGGGGC---ACCACCACUACCCGGCGGUCGUCCCGGAGGA---UCUCUUCCCCCA-CCAAUG .(((((..(((.(((...((.(((...))))).(((((.....)))))...))))))(((((---(((.((.......)).))).)))))((.(((---.....))).)).-.))))) ( -38.40, z-score = -0.98, R) >droSec1.super_37 372483 111 + 454039 CCAUUGCCACCAUCUACAGGUCGCCCAGCGCCAGCGAUUCCGAAUCGCCCAGGAGGUGGCGC---ACCACCACUACCCGGCGGUCGUCCCGGAGGA---UCUCUUCCCCCA-CCAAUG .....((((((.(((...((.(((...))))).(((((.....)))))...)))))))))((---(((.((.......)).))).))...((.(((---.....))).)).-...... ( -36.30, z-score = -1.18, R) >droYak2.chrX 9987289 111 + 21770863 CCAUUGCCACCAGCUACAGGUCGCCCAGCGCCAGCGAUUCCGAAUCGCCCAGGAGGUGGGGC---ACCACCACUACCCGGCGGUCGUCCCGGAGGA---GCUCUUCCCCCA-CCAAUG .(((((((.((.......((.(((...))))).(((((.....)))))...)).)).(((((---(((.((.......)).))).)))))(((((.---...)))))....-.))))) ( -37.80, z-score = -0.25, R) >droEre2.scaffold_4690 7497366 111 - 18748788 CCAUUGCCACCAGCAACAGGGCGCCCAGCGCCAGCGAUUCCGAAUCGCCCAGGAGGUGGGGC---ACCACCACUACCCGGCGGUCGUCCCGGAGGA---GCUCUUCCCCCA-CCAAUG ((.((((.....))))...(((((...))))).(((((.....)))))...)).((((((((---(((.((.......)).))).))...(((((.---...)))))))))-)).... ( -41.60, z-score = -0.86, R) >droAna3.scaffold_13417 5145304 111 + 6960332 CCGUUGCCUCCAUCUACAGGUCGCCCAGCUCCAGCGAUUCCGAAUCGUCCAGGAGGUGGAGC---CCCACCUCUUCCUGGAAGUCGCCCAGUAGGU---GCGUUGCCACCA-CCAAUA ..((.((..((((((((.((((((.........)))))).(((....((((((((((((...---.)))))...)))))))..)))....))))))---).)..)).))..-...... ( -39.40, z-score = -2.71, R) >dp4.Unknown_group_88 14509 111 - 30850 CCAUUACCACCGGCAACUGGGCGCCCAGCGCCAGCUAUUCCCAAUCGCCCAGGAGGUGGUGC---ACCACCACUACCUGGUGGUCGACCAGGAGGC---GCAUUGCCUCCU-CCAAUG .((((((((((((...(((((((...(((....))).........)))))))..(((((...---.))))).....)))))))).....(((((((---.....)))))))-..)))) ( -48.00, z-score = -2.93, R) >droPer1.super_141 49305 111 + 87512 CCAUUACCACCGGCAACUGGGCGCCCAGCGCCAGCUAUUCCCAAUCGCCCAGGAGGUGGUGC---ACCACCACUACCUGGUGGUCGACCAGGAGGC---GCAUUGCCUCCU-CCAAUG .((((((((((((...(((((((...(((....))).........)))))))..(((((...---.))))).....)))))))).....(((((((---.....)))))))-..)))) ( -48.00, z-score = -2.93, R) >droWil1.scaffold_181096 6530771 111 - 12416693 CCAUUGCCACCGGCAACUGGUCGCCCAGCACCAGCAAUUCCAAGUCGUCCUGGAGGCGGAGC---ACCUCCACUACCUGGCGGUCGGCCAGGUGGU---GCUCUGCCGCCC-CCACUG ...(((((...))))).(((..((((((.((..((........)).)).)))).((((((((---(((......(((((((.....))))))))))---))))))))))..-)))... ( -49.90, z-score = -2.32, R) >droGri2.scaffold_15203 5321952 111 - 11997470 CCAUUGCCACCGGCAACUGGACGCCCAGCGCCAGCAAUACCCAGCCGCCCAGGAGGUGGUGC---ACCACCGCUACCAGGCGGUCGGCCAGGCGGC---GCCCUUCCACCA-CCAUUG ((((((((...))))).))).......(((((.((.....((.((((((..((.(((((((.---..))))))).)).)))))).))....)))))---))..........-...... ( -46.60, z-score = -1.60, R) >anoGam1.chr2R 31534300 117 - 62725911 CCACCAGCACCAGCUAGUGCUAGGCCGGCUCCGGCAAUACCGAACCGACCUGGAGGUGGUGCCGCACCACCCCUGCCCGGUGGCCGUCCAGGUGGCCAGGCACUACCGGCA-CCAUUG ......((....(.(((((((.((((((...(((.....)))..)).((((((((((((((...)))))))...(((....)))..))))))))))).))))))).).)).-...... ( -51.60, z-score = -0.89, R) >apiMel3.Group3 10461153 112 - 12341916 CAGCCACCCCCUCUUUCUAGUUCUCGAGCUCCACC---UCCAGUUCCUACGGGUGGCCGACC---AGCGCCGGCUAUUCCUAACCGACCUGGACCUGGUGGACCUCCGCCAGCCCGUG ..(((((((...((....)).....(((((.....---...)))))....)))))))((.((---((.(.(((.((....)).))).)))))..(((((((....)))))))..)).. ( -37.10, z-score = -1.61, R) >consensus CCAUUGCCACCAGCUACAGGUCGCCCAGCGCCAGCGAUUCCGAAUCGCCCAGGAGGUGGGGC___ACCACCACUACCCGGCGGUCGUCCAGGAGGA___GCUCUUCCCCCA_CCAAUG ..((((((.((.......((....)).((....))................)).(((((.......))))).......))))))......((.(((........))).))........ (-19.04 = -19.48 + 0.43)
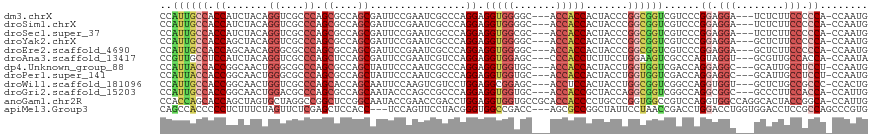
| Location | 15,796,246 – 15,796,357 |
|---|---|
| Length | 111 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 75.70 |
| Shannon entropy | 0.53857 |
| G+C content | 0.66231 |
| Mean single sequence MFE | -53.06 |
| Consensus MFE | -24.79 |
| Energy contribution | -25.54 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.59 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.698612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
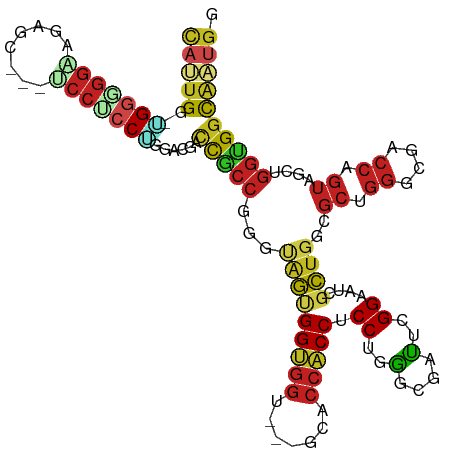
>dm3.chrX 15796246 111 - 22422827 CAUUGG-UGGGGGAAGAGA---UCCUCCGGGACGACCGCCGGGUAGUGGUGGU---GCCCCACCUCCUGGGCGAUUCGGAAUCGCUGGCGCUGGGCGACCUGUAGAUGGUGGCAAUGG (((((.-.((((((.....---))))))((.((.(((((......))))).))---.))(((((..(((((((((.....))))))(((((...))).))..)))..)))))))))). ( -47.80, z-score = -0.44, R) >droSim1.chrX 12215142 111 - 17042790 CAUUGG-UGGGGGAAGAGA---UCCUCCGGGACGACCGCCGGGUAGUGGUGGU---GCCCCACCUCCUGGGCGAUUCGGAAUCGCUGGCGCUGGGCGACCUGUAGAUGGUGGCAAUGG (((((.-.((((((.....---))))))((.((.(((((......))))).))---.))(((((..(((((((((.....))))))(((((...))).))..)))..)))))))))). ( -47.80, z-score = -0.44, R) >droSec1.super_37 372483 111 - 454039 CAUUGG-UGGGGGAAGAGA---UCCUCCGGGACGACCGCCGGGUAGUGGUGGU---GCGCCACCUCCUGGGCGAUUCGGAAUCGCUGGCGCUGGGCGACCUGUAGAUGGUGGCAAUGG (((((.-(((((((.....---)))))))......((((((..((..(((.((---((((((((....))(((((.....)))))))))))...)).)))..))..))))))))))). ( -47.50, z-score = -0.67, R) >droYak2.chrX 9987289 111 - 21770863 CAUUGG-UGGGGGAAGAGC---UCCUCCGGGACGACCGCCGGGUAGUGGUGGU---GCCCCACCUCCUGGGCGAUUCGGAAUCGCUGGCGCUGGGCGACCUGUAGCUGGUGGCAAUGG (((((.-.((((((.....---))))))((.((.(((((......))))).))---.))(((((.....((((((.....))))))(((((.((....)).)).))))))))))))). ( -48.90, z-score = -0.00, R) >droEre2.scaffold_4690 7497366 111 + 18748788 CAUUGG-UGGGGGAAGAGC---UCCUCCGGGACGACCGCCGGGUAGUGGUGGU---GCCCCACCUCCUGGGCGAUUCGGAAUCGCUGGCGCUGGGCGCCCUGUUGCUGGUGGCAAUGG (((((.-(((((((.....---)))))))......(((((((.(((..(.(((---((((...(.((..((((((.....)))))))).)..))))))))..))))))))))))))). ( -52.60, z-score = -0.59, R) >droAna3.scaffold_13417 5145304 111 - 6960332 UAUUGG-UGGUGGCAACGC---ACCUACUGGGCGACUUCCAGGAAGAGGUGGG---GCUCCACCUCCUGGACGAUUCGGAAUCGCUGGAGCUGGGCGACCUGUAGAUGGAGGCAACGG ((((..-.(((.((..(((---.((....)))))....((((...(((((((.---...)))))))(..(.((((.....)))))..)..)))))).)))....))))..(....).. ( -40.50, z-score = -0.21, R) >dp4.Unknown_group_88 14509 111 + 30850 CAUUGG-AGGAGGCAAUGC---GCCUCCUGGUCGACCACCAGGUAGUGGUGGU---GCACCACCUCCUGGGCGAUUGGGAAUAGCUGGCGCUGGGCGCCCAGUUGCCGGUGGUAAUGG ((((..-(((((((.....---))))))).....(((((((((..((((((..---.))))))..))).((((((((((..((((....))))....)))))))))))))))))))). ( -60.30, z-score = -3.66, R) >droPer1.super_141 49305 111 - 87512 CAUUGG-AGGAGGCAAUGC---GCCUCCUGGUCGACCACCAGGUAGUGGUGGU---GCACCACCUCCUGGGCGAUUGGGAAUAGCUGGCGCUGGGCGCCCAGUUGCCGGUGGUAAUGG ((((..-(((((((.....---))))))).....(((((((((..((((((..---.))))))..))).((((((((((..((((....))))....)))))))))))))))))))). ( -60.30, z-score = -3.66, R) >droWil1.scaffold_181096 6530771 111 + 12416693 CAGUGG-GGGCGGCAGAGC---ACCACCUGGCCGACCGCCAGGUAGUGGAGGU---GCUCCGCCUCCAGGACGACUUGGAAUUGCUGGUGCUGGGCGACCAGUUGCCGGUGGCAAUGG ...(((-((((((...(((---((((((((((.....)))))))......)))---)))))))))))).............(..((((((((((....)))).))))))..)...... ( -55.80, z-score = -1.82, R) >droGri2.scaffold_15203 5321952 111 + 11997470 CAAUGG-UGGUGGAAGGGC---GCCGCCUGGCCGACCGCCUGGUAGCGGUGGU---GCACCACCUCCUGGGCGGCUGGGUAUUGCUGGCGCUGGGCGUCCAGUUGCCGGUGGCAAUGG ....((-(((((......)---))))))..(((.((((((..(..(.(((((.---...))))).))..))))).).))).(..((((((((((....)))).))))))..)...... ( -56.00, z-score = -0.47, R) >anoGam1.chr2R 31534300 117 + 62725911 CAAUGG-UGCCGGUAGUGCCUGGCCACCUGGACGGCCACCGGGCAGGGGUGGUGCGGCACCACCUCCAGGUCGGUUCGGUAUUGCCGGAGCCGGCCUAGCACUAGCUGGUGCUGGUGG ......-..(((((((((((..((((((((((..(((....)))...(((((((...)))))))))))))).)))..))))))))))).(((((((((((....))))).)))))).. ( -69.40, z-score = -3.13, R) >apiMel3.Group3 10461153 112 + 12341916 CACGGGCUGGCGGAGGUCCACCAGGUCCAGGUCGGUUAGGAAUAGCCGGCGCU---GGUCGGCCACCCGUAGGAACUGGA---GGUGGAGCUCGAGAACUAGAAAGAGGGGGUGGCUG ......((((.((....)).))))(.((((((((((((....)))))))).))---)).)((((((((.......((.((---(......))).))..((......)).)))))))). ( -49.80, z-score = -1.98, R) >consensus CAUUGG_UGGGGGAAGAGC___UCCUCCUGGACGACCGCCGGGUAGUGGUGGU___GCACCACCUCCUGGGCGAUUCGGAAUCGCUGGCGCUGGGCGACCAGUAGCUGGUGGCAAUGG (((((...((((((........)))))).......(((((...(((((((((.......))))).((..(....)..))....))))..((.((....)).))....)))))))))). (-24.79 = -25.54 + 0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:47:54 2011