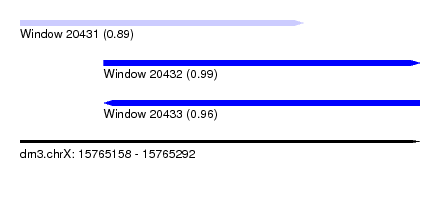
| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,765,158 – 15,765,292 |
| Length | 134 |
| Max. P | 0.991112 |

| Location | 15,765,158 – 15,765,253 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 88.87 |
| Shannon entropy | 0.18795 |
| G+C content | 0.28887 |
| Mean single sequence MFE | -18.84 |
| Consensus MFE | -15.52 |
| Energy contribution | -15.48 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.893402 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
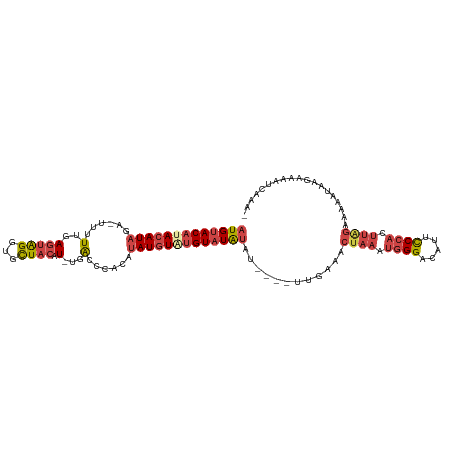
>dm3.chrX 15765158 95 + 22422827 GCAAAUGCUUGCUGUUACAGCUUUAAAAUGUGAUUUUCUUAUUUUU-CUAAGUGGGAAUGUCCCAUUUAGUUUCAA----AUAUAUACAUACAUAUGUGG ((((....)))).((((((.........)))))).....(((((..-(((((((((.....)))))))))....))----)))..(((((....))))). ( -19.90, z-score = -1.62, R) >droEre2.scaffold_4690 7465731 93 - 18748788 ---UUUAUGUGUUGCAACAGCUUUAAAAUUUGAGUUUCUUUUUUUUUCUAAGUGGGAAUGUCCCAUUUAGUUCCAU----AUAUAUACAUACAUAUGUGG ---..((((((..((...(((((........)))))............((((((((.....))))))))))..)))----)))..(((((....))))). ( -20.30, z-score = -2.41, R) >droYak2.chrX 9956731 95 + 21770863 ---UUUAUUUGUUGCAACAGCUUUAAA-UUCGAUUUUCUUAUUUUU-CCAAGUGGGAAUGUCCCAUUUAGUUCCAUCUAUAUACAUACAUACAUAUGUGG ---...(((((..((....))..))))-).................-..(((((((.....))))))).............((((((......)))))). ( -14.50, z-score = -0.84, R) >droSec1.super_37 341944 92 + 454039 ---UUUAUUUGCUGCAACAGCUUUAAAAUUUGAUUUUCUUAUUUUU-CUAAGUGGGAAUGUCCCAUUUAGUUUCAA----AUAUAUACAUACAUAUGUGG ---((((...((((...))))..))))((((((.............-(((((((((.....)))))))))..))))----))...(((((....))))). ( -19.76, z-score = -2.74, R) >droSim1.chrX 12195405 92 + 17042790 ---UUUAUUUGCUGCAACAGCUUUAAAAUUUGAUUUUCUUAUUUUU-CUAAGUGGGAAUGUCCCAUUUAGUUUCAA----AUAUAUACAUACAUAUGUGG ---((((...((((...))))..))))((((((.............-(((((((((.....)))))))))..))))----))...(((((....))))). ( -19.76, z-score = -2.74, R) >consensus ___UUUAUUUGCUGCAACAGCUUUAAAAUUUGAUUUUCUUAUUUUU_CUAAGUGGGAAUGUCCCAUUUAGUUUCAA____AUAUAUACAUACAUAUGUGG ..........((((...))))..........................(((((((((.....)))))))))...............(((((....))))). (-15.52 = -15.48 + -0.04)
| Location | 15,765,186 – 15,765,292 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 79.66 |
| Shannon entropy | 0.38484 |
| G+C content | 0.31408 |
| Mean single sequence MFE | -24.49 |
| Consensus MFE | -16.42 |
| Energy contribution | -18.53 |
| Covariance contribution | 2.11 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.991112 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
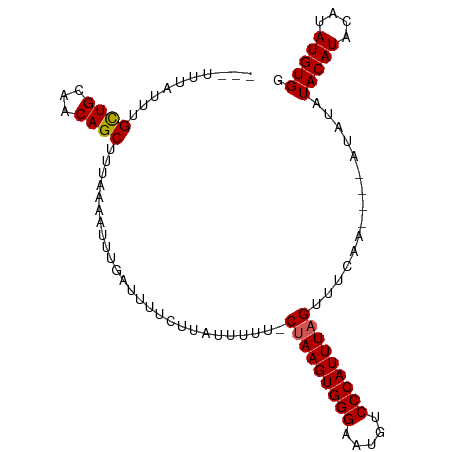
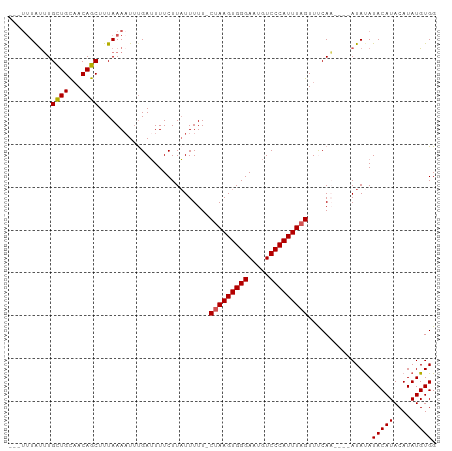
>dm3.chrX 15765186 106 + 22422827 -UGUGAUUUUCUUAUUUUUCUAAGUGGGAAUGUCCCAUUUAGUUUCAA----AUAUAUACAUACAUAUGUGGGUCA-AUGUAGCACCUACUCAAAAA-UCUAUGUAUGUACAU -.(((.......(((((..(((((((((.....)))))))))....))----)))..((((((((((.(((((((.-.....).)))))).......-..))))))))))))) ( -28.20, z-score = -3.47, R) >droEre2.scaffold_4690 7465756 108 - 18748788 UUUGAGUUUCUUUUUUUUUCUAAGUGGGAAUGUCCCAUUUAGUUCCAU----AUAUAUACAUACAUAUGUGGGUCA-AUGUAGCACCUACUCAAAAAAUCUAUGUGUGUACAU ...................(((((((((.....)))))))))......----.....((((((((((.(((((((.-.....).))))))..........))))))))))... ( -27.10, z-score = -2.89, R) >droYak2.chrX 9956755 111 + 21770863 -UUCGAUUUUCUUAUUUUUCCAAGUGGGAAUGUCCCAUUUAGUUCCAUCUAUAUACAUACAUACAUAUGUGGGUCA-AUGUAGCACCUACUCAAAAAAUCUAUGUGUGUACAU -....................(((((((.....))))))).............((((((((((.....(((((((.-.....).))))))..........))))))))))... ( -23.66, z-score = -1.81, R) >droSec1.super_37 341969 106 + 454039 -UUUGAUUUUCUUAUUUUUCUAAGUGGGAAUGUCCCAUUUAGUUUCAA----AUAUAUACAUACAUAUGUGGGUCA-AUGUAGCAUCUAUUCAAAAA-UCUAUGUAUGUACAU -...........(((((..(((((((((.....)))))))))....))----)))......(((((((((((((..-..((((...))))......)-))))))))))))... ( -24.70, z-score = -3.09, R) >droSim1.chrX 12195430 106 + 17042790 -UUUGAUUUUCUUAUUUUUCUAAGUGGGAAUGUCCCAUUUAGUUUCAA----AUAUAUACAUACAUAUGUGGGUCA-AUGUAGCACCUACUCAAAAA-UCUAUGUAUGUACAU -...........(((((..(((((((((.....)))))))))....))----)))..((((((((((.(((((((.-.....).)))))).......-..))))))))))... ( -28.00, z-score = -3.86, R) >droVir3.scaffold_13042 2695697 95 - 5191987 -----AAACACUUAUUCUU----AAGGAAAGGUCC---UUGGUCCUAG----GUAUAUACACAUAUUUGUAUGCCAUAUGUAA-GCCCCCUCAAAAAUCGCAU-UAAGUUCAG -----....(((((...((----(((((....)))---)))).....(----(((((((........))))))))........-...................-))))).... ( -15.30, z-score = -0.28, R) >consensus _UUUGAUUUUCUUAUUUUUCUAAGUGGGAAUGUCCCAUUUAGUUCCAA____AUAUAUACAUACAUAUGUGGGUCA_AUGUAGCACCUACUCAAAAA_UCUAUGUAUGUACAU ...................(((((((((.....)))))))))...............((((((((((.((((((..........))))))..........))))))))))... (-16.42 = -18.53 + 2.11)
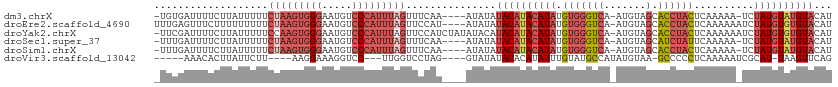
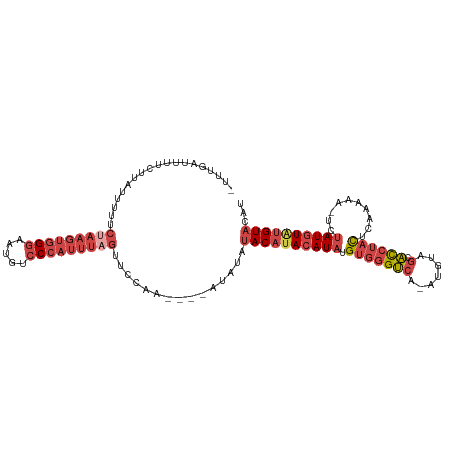
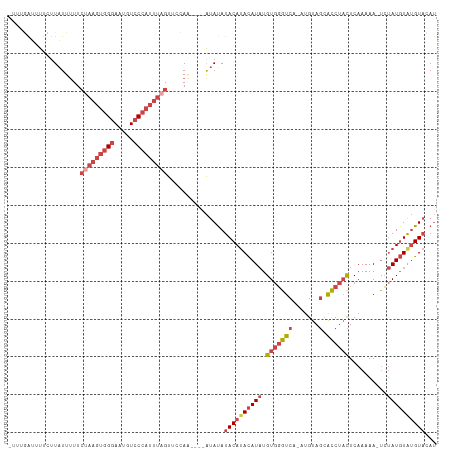
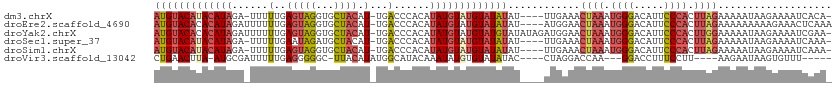
| Location | 15,765,186 – 15,765,292 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 79.66 |
| Shannon entropy | 0.38484 |
| G+C content | 0.31408 |
| Mean single sequence MFE | -23.70 |
| Consensus MFE | -14.48 |
| Energy contribution | -16.23 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.961077 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15765186 106 - 22422827 AUGUACAUACAUAGA-UUUUUGAGUAGGUGCUACAU-UGACCCACAUAUGUAUGUAUAUAU----UUGAAACUAAAUGGGACAUUCCCACUUAGAAAAAUAAGAAAAUCACA- (((((((((((((..-...(..(((((...)))).)-..)......)))))))))))))..----......((((.((((.....)))).))))..................- ( -25.50, z-score = -3.01, R) >droEre2.scaffold_4690 7465756 108 + 18748788 AUGUACACACAUAGAUUUUUUGAGUAGGUGCUACAU-UGACCCACAUAUGUAUGUAUAUAU----AUGGAACUAAAUGGGACAUUCCCACUUAGAAAAAAAAAGAAACUCAAA ((((....))))......(((((((.((..(.....-.)..)).((((((((....)))))----)))...((((.((((.....)))).))))............))))))) ( -23.20, z-score = -2.02, R) >droYak2.chrX 9956755 111 - 21770863 AUGUACACACAUAGAUUUUUUGAGUAGGUGCUACAU-UGACCCACAUAUGUAUGUAUGUAUAUAGAUGGAACUAAAUGGGACAUUCCCACUUGGAAAAAUAAGAAAAUCGAA- ((((....)))).((((((((..((((...))))..-....(((..(((((((....)))))))..)))..((((.((((.....)))).)))).......))))))))...- ( -24.80, z-score = -1.50, R) >droSec1.super_37 341969 106 - 454039 AUGUACAUACAUAGA-UUUUUGAAUAGAUGCUACAU-UGACCCACAUAUGUAUGUAUAUAU----UUGAAACUAAAUGGGACAUUCCCACUUAGAAAAAUAAGAAAAUCAAA- ((((....)))).((-((((((((((.(((((((((-((.....)).))))).)))).)))----))....((((.((((.....)))).))))........)))))))...- ( -22.80, z-score = -3.37, R) >droSim1.chrX 12195430 106 - 17042790 AUGUACAUACAUAGA-UUUUUGAGUAGGUGCUACAU-UGACCCACAUAUGUAUGUAUAUAU----UUGAAACUAAAUGGGACAUUCCCACUUAGAAAAAUAAGAAAAUCAAA- (((((((((((((..-...(..(((((...)))).)-..)......)))))))))))))..----......((((.((((.....)))).))))..................- ( -25.50, z-score = -3.19, R) >droVir3.scaffold_13042 2695697 95 + 5191987 CUGAACUUA-AUGCGAUUUUUGAGGGGGC-UUACAUAUGGCAUACAAAUAUGUGUAUAUAC----CUAGGACCAA---GGACCUUUCCUU----AAGAAUAAGUGUUU----- ........(-((((.(((((((((((((.-.((((((((.........))))))))....)----))(((.(...---.).)))..))))----))))))..))))).----- ( -20.40, z-score = -0.65, R) >consensus AUGUACAUACAUAGA_UUUUUGAGUAGGUGCUACAU_UGACCCACAUAUGUAUGUAUAUAU____UUGAAACUAAAUGGGACAUUCCCACUUAGAAAAAUAAGAAAAUCAAA_ (((((((((((((..........((((...))))............)))))))))))))............((((.((((.....)))).))))................... (-14.48 = -16.23 + 1.75)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:47:48 2011