
| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,763,177 – 15,763,294 |
| Length | 117 |
| Max. P | 0.559177 |

| Location | 15,763,177 – 15,763,294 |
|---|---|
| Length | 117 |
| Sequences | 15 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 77.62 |
| Shannon entropy | 0.51495 |
| G+C content | 0.50883 |
| Mean single sequence MFE | -37.18 |
| Consensus MFE | -16.86 |
| Energy contribution | -16.16 |
| Covariance contribution | -0.70 |
| Combinations/Pair | 1.65 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.559177 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
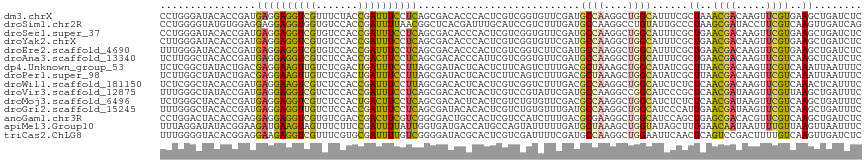
>dm3.chrX 15763177 117 + 22422827 CCUGGGAUACACCGAUGAGGAGGUCGUUUCUACCGAUUUCCUCAGCGACACCCACUCGUCGGUGUUCGAUGCCAAGGCUGGCAUUUCGCUAAACGACAAGUUCGUGAAGCUGAUCUC ...(((((((((((.(((((((((((.......)))))))))))((((.......))))))))))..(((((((....)))))))..(((..((((.....))))..)))..))))) ( -43.10, z-score = -2.54, R) >droSim1.chr2R 8013834 117 - 19596830 CCUGGGGUAUGUGGAGGAGGAGGUGGUGUCCACCGAUUUUAACGGCUCACGAUUUGCAUCCGUCUUUGAUGCCAAGGCCUGUAUUGCCCUAAGCGAUACCUUCGUCAAGUUGAUCAG .((((...((...((.((((.(((((...))))).........((((.....((.(((((.......))))).)))))).(((((((.....))))))))))).))..))...)))) ( -32.90, z-score = 0.66, R) >droSec1.super_37 339933 117 + 454039 CCUGGGAUACACCGAUGAGGAGGUCGUGUCCACCGAUUUCCUCAGCGACACCCACUCGUCGGUGUUCGAUGCCAAGGCUGGCAUUUCGCUGAACGACAAGUUCGUGAAGCUGAUCUC ...(((((((((((.(((((((((((.(....))))))))))))((((.......))))))))))..(((((((....)))))))..(((..((((.....))))..)))..))))) ( -44.80, z-score = -2.10, R) >droYak2.chrX 9954611 117 + 21770863 CUUGGGAUACACCGAUGAGGAGGUCGUGUCCACCGAUUUCCUCAGCGACACCCACUCGUCGGUGUUCGAUGCCAAGGCUGGCAUUUCGCUGAACGACAAGUUCGUGAAGCUGAUCUC ...(((((((((((.(((((((((((.(....))))))))))))((((.......))))))))))..(((((((....)))))))..(((..((((.....))))..)))..))))) ( -44.80, z-score = -2.16, R) >droEre2.scaffold_4690 7463669 117 - 18748788 UUUGGGAUACACCGAUGAGGAGGUCGUGUCCACCGAUUUCCUCAGCGACACCCACUCGUCGGUCUUCGAUGCCAAGGCUGGCAUUUCGCUGAACGACAAGUUCGUGAAGCUGAUCUC .((((......)))).((((((((((.(....)))))))))))(((..(((..(((.((((.((..((((((((....)))))..)))..)).)))).)))..)))..)))...... ( -44.60, z-score = -2.35, R) >droAna3.scaffold_13340 16114294 117 - 23697760 UCUUGGCUACACCGAUGAGGAGGUCGUCUCCACCGACUUCCUCAGCGACACCCAUUCGUCGGUGUUCGAUGCCAAGGCUGGCAUUUCGCUGAACGACAAGUUCGUCAAGCUCAUCUC .((((((.((((((.(((((((((((.......)))))))))))((((.......)))))))))).....))))))((((((.....)))((((.....))))....)))....... ( -46.10, z-score = -3.55, R) >dp4.Unknown_group_53 33876 117 - 37457 UCUCGGCUAUACUGACGAGGAAGUUGUCUCGACUGAUUUCCUUAGCGAUACUCACUCUUCAGUCUUUGACGCUAAAGCUGGCAUAUCGCUUAACGACAAGUUCGUCAAAUUAAUUUC ............(((((((((((((((....)).)))))))..(((((((...........(((...)))((((....)))).)))))))...........)))))).......... ( -26.90, z-score = -0.72, R) >droPer1.super_98 66573 117 - 160379 UCUUGGCUAUACUGACGAGGAAGUUGUCUCGACUGAUUUCCUUAGCGAUACUCACUCUUCAGUCUUUGACGCUAAAGCUGGCAUAUCGCUUAACGACAAGUUCGUCAAAUUAAUUUC ............(((((((((((((((....)).)))))))..(((((((...........(((...)))((((....)))).)))))))...........)))))).......... ( -26.90, z-score = -0.83, R) >droWil1.scaffold_181150 2811765 117 + 4952429 UCUCGGCUACACCGAUGAGGAAGUCGUCUCCACCGAUUUCCUUAGCGACACUCACUCGUCGGUCUUUGACGCCAAGGCUGGCAUCUCUCUCAACGACAAGUUCGUCAAACUCAUUUC ..((((.....))))(((((((((((.......)))))))))))..(((....(((.(((((((...)))((((....))))...........)))).)))..)))........... ( -33.90, z-score = -2.13, R) >droVir3.scaffold_12875 8989492 117 - 20611582 UUUGGGCUAUACCGAUGAGGAGGUCGUCUCCACCGAUUUCCUCAGCGACACUCACUCGUCCGUAUUCGAUGCCAAGGCCGGCAUCCCGCUCAACGAUAAGUUCGUUAAGCUGAUUUC ...((...((((.((((((((((((((.................))))).))).)))))).))))..((((((......))))))))(((.(((((.....))))).)))....... ( -40.03, z-score = -2.62, R) >droMoj3.scaffold_6496 15250971 117 + 26866924 UCUGGGCUACACCGAUGAGGAGGUCGUCUCCACUGACUUCCUCAGCGACACUCACUCGUCUGUGUUCGACGCCAAGGCUGGCAUCUCUCUCAACGAUAAGUUCGUCAAGCUGAUUUC ....((((((((.(((((((((((((......((((.....)))))))).))).)))))).))))..(((((((....))))............((.....))))).))))...... ( -36.60, z-score = -0.98, R) >droGri2.scaffold_15245 6602792 117 + 18325388 UUUGGGCUACACCGAUGAGGAGGUUGUCUCCACCGAUUUCCUCAGCGAUACACACUCGUCUGUGUUUGAUGCCAAGGCUGGCAUCCCAUUGAACGAUAAGUUCGUCAAGCUGAUUUC ..(.((((((((.((((((((((..(((......)))..))))...(.....).)))))).))))..(((((((....))))))).....((((.....))))....)))).).... ( -37.00, z-score = -1.65, R) >anoGam1.chr3R 37843862 117 + 53272125 CCUGGACUACACCGAGGAGGAGGUCGUGUCGACCGACUUCGUCGGCGACUGCCACUCGUCCAUCUUUGACGCGAAGGCUGGCAUCCAGCUGAGCGACACGUUCGUCAAGCUGAUCUC ...(((...(.((((((((..(((((...)))))..)))).)))).)..(((((...(((.......)))((....))))))))))((((..((((.....))))..))))...... ( -42.90, z-score = -0.28, R) >apiMel3.Group10 9158520 117 - 11440700 UUUAGGAUAUACGGAAGAUGAAGUAGUUUCUUCCGAUUUUAUUGGUGAUGACCAUGCCAGUAUUUUUGAUGCUAAAGCUGGUAUAGCUUUGAACAAUAAUUUUGUUAAGUUAAUUUC ...........(((((((..........))))))).......((((....))))(((((((.(((........))))))))))((((((..(((((.....)))))))))))..... ( -26.20, z-score = -1.80, R) >triCas2.ChLG8 2303111 117 + 15773733 UUUGGGGUACACGGAGGAAGAGGUCGUUUCGUGCGAUUUUGUCGGGGAUACGCACUCGUCGAUUUUCGAUGCCAAGGCUGGAAUUCAACUCAGUCCGACUUUUGUCAAGUUGAUCUC .((((.((.(((((((((.....)).)))))))(((..(((.((((........)))).)))...))))).))))((((((........))))))((((((.....))))))..... ( -31.00, z-score = 0.79, R) >consensus UCUGGGCUACACCGAUGAGGAGGUCGUCUCCACCGAUUUCCUCAGCGACACCCACUCGUCGGUCUUCGAUGCCAAGGCUGGCAUUUCGCUGAACGACAAGUUCGUCAAGCUGAUCUC ................((((((((((.......))))))))))...........................((((....))))......((..((((.....))))..))........ (-16.86 = -16.16 + -0.70)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:47:46 2011