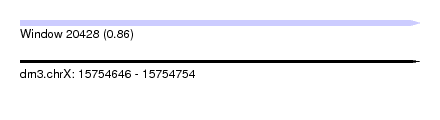
| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,754,646 – 15,754,754 |
| Length | 108 |
| Max. P | 0.857950 |

| Location | 15,754,646 – 15,754,754 |
|---|---|
| Length | 108 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 64.07 |
| Shannon entropy | 0.78080 |
| G+C content | 0.51005 |
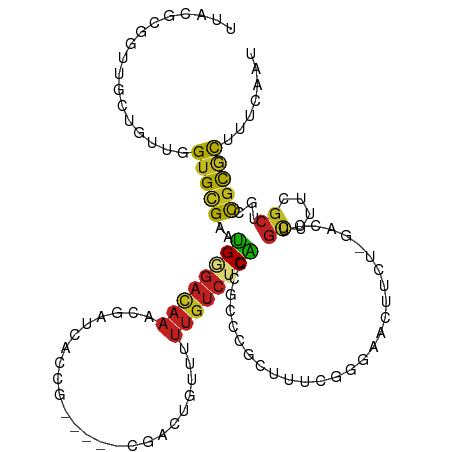
| Mean single sequence MFE | -30.06 |
| Consensus MFE | -10.39 |
| Energy contribution | -10.14 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.857950 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
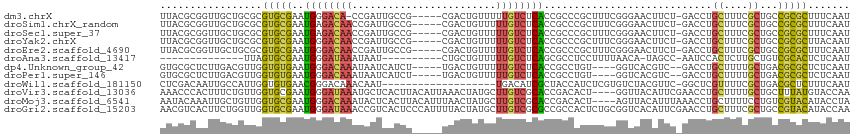
>dm3.chrX 15754646 108 + 22422827 UUACGCGGUUGCUGCGCGUGCGAAUGGGACA-CCGAUUGCCG-----CGACUGUUUUUGUCUCACCGCCCGCUUUCGGGAACUUCU-GACCUGCUUUCGCUGCCGCGCUUUCAAU ...((((((.((...(((.((((.(((....-))).))))))-----)((..((....(((......((((....)))).......-)))..))..)))).))))))........ ( -37.32, z-score = -1.33, R) >droSim1.chrX_random 33828 109 + 5698898 UUACGCGGUUGCUGCGCGUGCGAAUGAGACAACCGAUUGCCG-----CGACUGUUUUUGUCUCACCGCCCGCUUUCGGGAACUUCU-GACCUGCUUUCGCUGCCGCGCUUUCAAU ...((((((.((.(((((.(((..((((((((..(((.(...-----...).))).)))))))).))).)))..(((((....)))-))...))....)).))))))........ ( -34.70, z-score = -1.04, R) >droSec1.super_37 331277 109 + 454039 UUACGCGGUUGCUGCGCGUGCGAAUGAGACAACCGAUUGCCG-----CGACUGUUUUUGUCUCACCGCCCGCUUUCGGGAACUUCU-GACCUGCUUUCGCUGCCGCGCUUUCAAU ...((((((.((.(((((.(((..((((((((..(((.(...-----...).))).)))))))).))).)))..(((((....)))-))...))....)).))))))........ ( -34.70, z-score = -1.04, R) >droYak2.chrX 9946140 109 + 21770863 UUACGCGGUUGCUGCGCGUGCGAAUGGGACAACCGAUUGCCG-----CGACUGUUUUUGUCUCACCGCCCGCUUUCGGGAACUUCU-GACCUGCUUUCGCUGCCGCGCUUACAAU ...((((((.((...(((.((((.(((.....))).))))))-----)((..((....(((......((((....)))).......-)))..))..)))).))))))........ ( -37.32, z-score = -1.45, R) >droEre2.scaffold_4690 7455282 109 - 18748788 UUACGCGGUUGCUGCGCGUGCGAAUGGGACAACCGAUUGCCG-----CGACUGUUUUUGUCUCACCGCCCGCUUUCGGGAACUUCU-GACCUGCUUUCGCUGCCGCGCUUUCAAU ...((((((.((...(((.((((.(((.....))).))))))-----)((..((....(((......((((....)))).......-)))..))..)))).))))))........ ( -37.32, z-score = -1.35, R) >droAna3.scaffold_13417 4123686 89 + 6960332 --------------UUAGUGCGAAUGGGAUAAAUAAU----------CUGCUGUUUUUGUCUCAGCGCCUCCUUUUAACA-UAGCC-AAUCCACUCUUGCUGUCGCACUCUCAAU --------------..(((((((.(((((((((....----------........)))))))))................-((((.-...........)))))))))))...... ( -18.60, z-score = -1.43, R) >dp4.Unknown_group_42 5682 104 + 36417 GUGCGCUCUUGACGUUGGUGUGAAUGGGACAAAUAAUCAUCU-----UGACUGUUUUUGUCUCACCGCCUGU----GGUCACGUC--GACCUGCUUUUGCUGACGCGCUCUCAAU (((((.((.....((((..((((.(((((((((...((....-----.)).....)))))))))(((....)----))))))..)--)))..((....)).)))))))....... ( -31.10, z-score = -1.91, R) >droPer1.super_146 16091 104 + 80581 GUGCGCUCUUGACGUUGGUGUGAAUGGGACAAAUAAUCAUCU-----UGACUGUUUUUGUCUCACCGCCUGU----GGUCACGUC--GACCUGCUUUUGCUGACGCGCUCUCAAU (((((.((.....((((..((((.(((((((((...((....-----.)).....)))))))))(((....)----))))))..)--)))..((....)).)))))))....... ( -31.10, z-score = -1.91, R) >droWil1.scaffold_181150 2065114 95 - 4952429 CUCGACAAUUGCCAUUGGUGUGAACGGGACAAACAAU-------------------UGACAUCGCUACCAUCUCGUGUCUACGUUC-GGCUCGUUUUCGCUGACGCUCUUUCAAU ...((.((..((..(..(((.(((((((.(.(((...-------------------.((((.((.........))))))...))).-).))))))).)))..).))..))))... ( -19.80, z-score = 0.38, R) >droVir3.scaffold_13036 972973 111 + 1135500 AAACCCACUUUCUGUUGGUGCGAAUGGGAUAAAUGCUCACUUACAUUAAACUAUGCUUGUCGCACCGACACU----GGUUACAUUCGAACCUGCUUUUGCUGCUUUAUGUACCAA ............(((((((((((..((.(((((((........))))....))).))..))))))))))).(----(((.((((..(((.(.((....)).).))))))))))). ( -28.50, z-score = -3.12, R) >droMoj3.scaffold_6541 1612881 111 + 2543558 AAUACAAAUUGCUGUUGGUGCGAAUGGGACAAAUACUCACUUACAUUUAACUAUGCUUGUCGCACCGACACU----AGUUACAUUUAAACCUGCUUUUCCUGUCGUACAUACCUA ......(((((.(((((((((((.((((.......))))....(((......)))....))))))))))).)----))))................................... ( -22.10, z-score = -1.55, R) >droGri2.scaffold_15203 11035435 115 + 11997470 AACGUCACUUCUGGUUGGUGCGAAUGGGAUAAACCGUCACUCCCAUUUUACUAUGCUUGUCGCGCCGCCACUCUGCGGUCACAUUCGAACCUGCUUUCGCUGCCGUACAUACCAA ...........((((.(((((((((((((...........)))))).............)))))))))))...(((((.((....((((......)))).)))))))........ ( -28.21, z-score = -0.38, R) >consensus UUACGCGGUUGCUGUUGGUGCGAAUGGGACAAACGAUCACCG_____CGACUGUUUUUGUCUCACCGCCCGCUUUCGGGAACUUCU_GACCUGCUUUCGCUGCCGCGCUUUCAAU .................(((((..((((((((........................))))))))............................((....))...)))))....... (-10.39 = -10.14 + -0.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:47:44 2011