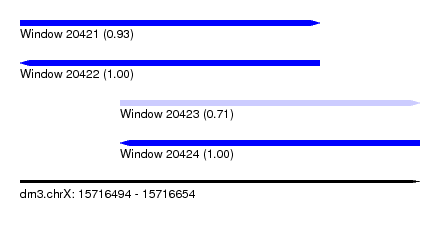
| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,716,494 – 15,716,654 |
| Length | 160 |
| Max. P | 0.999055 |

| Location | 15,716,494 – 15,716,614 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.36 |
| Shannon entropy | 0.20494 |
| G+C content | 0.43937 |
| Mean single sequence MFE | -36.95 |
| Consensus MFE | -23.65 |
| Energy contribution | -23.53 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.925146 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15716494 120 + 22422827 CACUAACGUAGACACCUCUUUGAAUGAGAUUUCGAAUCCUCGGAGGCUUUUGGGUGUCCAUAAGUACGCAACGAAGAAUAUGGCAUAUUCGAGAUAUUCUAUAGCAUACUUUUGGACACU ........((((..((((((((((......)))))......)))))..))))((((((((.(((((.((.....(((((((..(......)..)))))))...)).))))).)))))))) ( -32.50, z-score = -2.53, R) >droEre2.scaffold_4690 7422442 118 - 18748788 CACU--CGUAGACACCUCUUUGAAUGUGAUUUCGCGUCCUCGGAGGCUGUAAGGUGUCCGAAAGUAUGCAGUGAAGAACAUGGCAUGGUCAAGAUAUUCUAUAGCAUACUUUCAGACAGU ..((--..(((...(((((..(.(((((....))))).)..))))))))..)).((((.((((((((((.(((((...(.((((...)))).)...))).)).)))))))))).)))).. ( -37.50, z-score = -2.29, R) >droSec1.super_37 297444 120 + 454039 CACUAACGUAGACACCUCUUUGAAUGAGAUUUCGAAUCCUCGUAGGCUUUUGGGUGUCCGUGAGUACGCAACGAAGAAUAUGGCAUAGCCGCCAUAUUCUGUAGCAUACUUUUGGACACU ..(((.((..((......((((((......))))))))..))))).......((((((((.(((((.((.((..((((((((((......)))))))))))).)).))))).)))))))) ( -40.50, z-score = -3.79, R) >droSim1.chrU 78422 120 + 15797150 CACUAACGUAGCCACCUCUUGGAAUGAGAUUUCGAAUCCUCGCAGGCUUUUGGGUGUCCGUAAGUACGCAACGAAUAAUAUGGCAUAGCCGAGAUAUUCUGUAGCAUACUUUUGGACACU ..((((...((((.((....))...(((..........)))...)))).))))(((((((.(((((.((.(((((((.(.((((...)))).).))))).)).)).))))).))))))). ( -37.30, z-score = -2.78, R) >consensus CACUAACGUAGACACCUCUUUGAAUGAGAUUUCGAAUCCUCGGAGGCUUUUGGGUGUCCGUAAGUACGCAACGAAGAAUAUGGCAUAGCCGAGAUAUUCUAUAGCAUACUUUUGGACACU ..............(((((..(.((..(....)..)).)..)))))......((((((((.(((((.((.(((((.....((((...)))).....))).)).)).))))).)))))))) (-23.65 = -23.53 + -0.12)

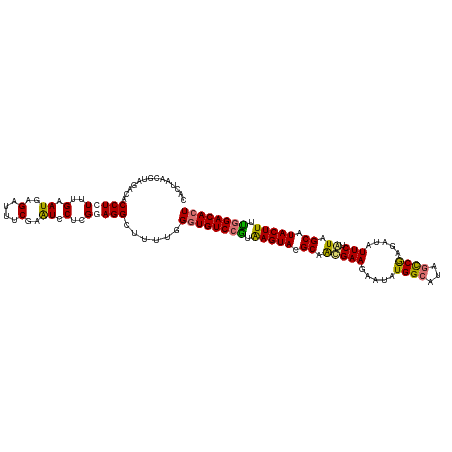
| Location | 15,716,494 – 15,716,614 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.36 |
| Shannon entropy | 0.20494 |
| G+C content | 0.43937 |
| Mean single sequence MFE | -37.80 |
| Consensus MFE | -26.60 |
| Energy contribution | -27.73 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.71 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.88 |
| SVM RNA-class probability | 0.996067 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15716494 120 - 22422827 AGUGUCCAAAAGUAUGCUAUAGAAUAUCUCGAAUAUGCCAUAUUCUUCGUUGCGUACUUAUGGACACCCAAAAGCCUCCGAGGAUUCGAAAUCUCAUUCAAAGAGGUGUCUACGUUAGUG .(((((((.((((((((...(((((((..(......)..))))))).....)))))))).)))))))......(((((...((((..(....)..))))...)))))............. ( -33.50, z-score = -3.42, R) >droEre2.scaffold_4690 7422442 118 + 18748788 ACUGUCUGAAAGUAUGCUAUAGAAUAUCUUGACCAUGCCAUGUUCUUCACUGCAUACUUUCGGACACCUUACAGCCUCCGAGGACGCGAAAUCACAUUCAAAGAGGUGUCUACG--AGUG (((....((((((((((...(((((((............))))))).....))))))))))(((((((((...((.((....)).))(((......)))...)))))))))...--))). ( -36.50, z-score = -3.36, R) >droSec1.super_37 297444 120 - 454039 AGUGUCCAAAAGUAUGCUACAGAAUAUGGCGGCUAUGCCAUAUUCUUCGUUGCGUACUCACGGACACCCAAAAGCCUACGAGGAUUCGAAAUCUCAUUCAAAGAGGUGUCUACGUUAGUG .((((((...(((((((.(((((((((((((....)))))))))))..)).)))))))...))))))......((..(((.((((.....(((((.......))))))))).)))..)). ( -43.80, z-score = -5.04, R) >droSim1.chrU 78422 120 - 15797150 AGUGUCCAAAAGUAUGCUACAGAAUAUCUCGGCUAUGCCAUAUUAUUCGUUGCGUACUUACGGACACCCAAAAGCCUGCGAGGAUUCGAAAUCUCAUUCCAAGAGGUGGCUACGUUAGUG .((((((..((((((((.((.(((((....(((...)))....))))))).))))))))..)))))).....((((..((......))..(((((.......)))))))))......... ( -37.40, z-score = -3.01, R) >consensus AGUGUCCAAAAGUAUGCUACAGAAUAUCUCGACUAUGCCAUAUUCUUCGUUGCGUACUUACGGACACCCAAAAGCCUCCGAGGAUUCGAAAUCUCAUUCAAAGAGGUGUCUACGUUAGUG .(((((((.((((((((...(((((((..((....))..))))))).....)))))))).)))))))......((..(((.((((.....(((((.......))))))))).)))..)). (-26.60 = -27.73 + 1.12)

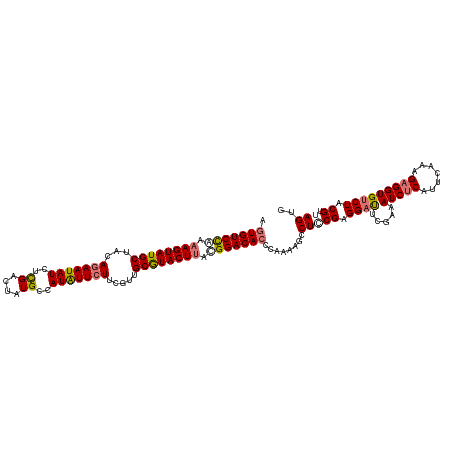
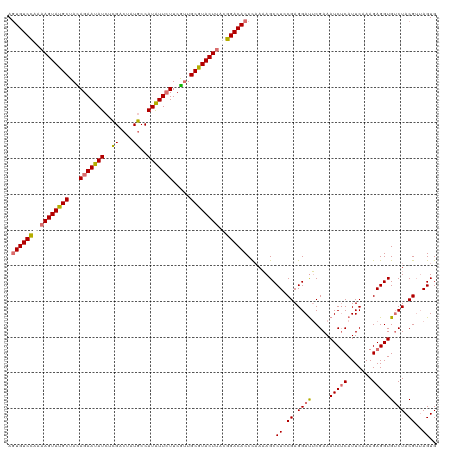
| Location | 15,716,534 – 15,716,654 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.92 |
| Shannon entropy | 0.21068 |
| G+C content | 0.44273 |
| Mean single sequence MFE | -35.88 |
| Consensus MFE | -25.59 |
| Energy contribution | -25.40 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.707465 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15716534 120 + 22422827 CGGAGGCUUUUGGGUGUCCAUAAGUACGCAACGAAGAAUAUGGCAUAUUCGAGAUAUUCUAUAGCAUACUUUUGGACACUUUGUUGAUGAAGGGAUUUUCGAUCUAUCGCACCCGUGUAU (((..((....(((((((((.(((((.((.....(((((((..(......)..)))))))...)).))))).))))))))).......((..(((((...))))).))))..)))..... ( -34.20, z-score = -2.05, R) >droEre2.scaffold_4690 7422480 117 - 18748788 CGGAGGCUGUAAGGUGUCCGAAAGUAUGCAGUGAAGAACAUGGCAUGGUCAAGAUAUUCUAUAGCAUACUUUCAGACAGUCUGUUGAA---GGGAUUUUCGAGCUAUCGCACCCAUGUAU .((..((.((((((((((.((((((((((.(((((...(.((((...)))).)...))).)).)))))))))).)))).))).(((((---(....))))))..))).))..))...... ( -32.60, z-score = -1.08, R) >droSec1.super_37 297484 117 + 454039 CGUAGGCUUUUGGGUGUCCGUGAGUACGCAACGAAGAAUAUGGCAUAGCCGCCAUAUUCUGUAGCAUACUUUUGGACACUUUGUUGAA---GGGAUUUUCGAUCUAUCGCACCCGUGUAU .(((.((....(((((((((.(((((.((.((..((((((((((......)))))))))))).)).))))).))))))))).......---(((.....(((....)))..))))).))) ( -41.70, z-score = -3.44, R) >droSim1.chrU 78462 116 + 15797150 CGCAGGCUUUUGGGUGUCCGUAAGUACGCAACGAAUAAUAUGGCAUAGCCGAGAUAUUCUGUAGCAUACUUUUGGACACUUUGUUGA----GGGAUUUUCGAUCUAUCGCACCCGUGUAU (((.((.....(((((((((.(((((.((.(((((((.(.((((...)))).).))))).)).)).))))).)))))))))(((.((----.(((((...))))).))))))).)))... ( -35.00, z-score = -1.78, R) >consensus CGGAGGCUUUUGGGUGUCCGUAAGUACGCAACGAAGAAUAUGGCAUAGCCGAGAUAUUCUAUAGCAUACUUUUGGACACUUUGUUGAA___GGGAUUUUCGAUCUAUCGCACCCGUGUAU ....(((....(((((((((.(((((.((.(((((.....((((...)))).....))).)).)).))))).))))))))).)))......(((.....(((....)))..)))...... (-25.59 = -25.40 + -0.19)

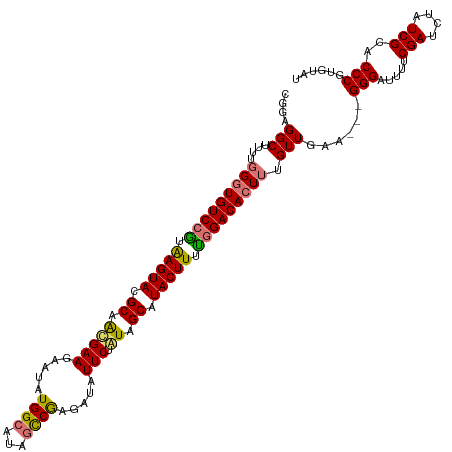
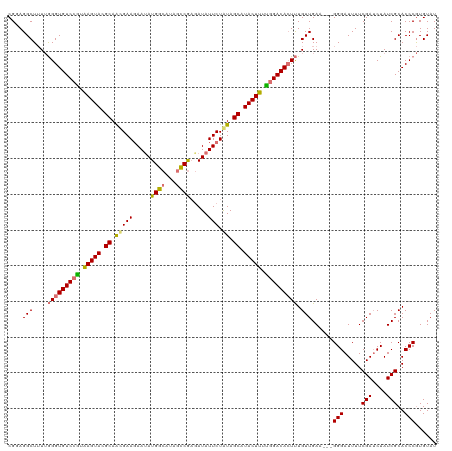
| Location | 15,716,534 – 15,716,654 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.92 |
| Shannon entropy | 0.21068 |
| G+C content | 0.44273 |
| Mean single sequence MFE | -35.88 |
| Consensus MFE | -27.50 |
| Energy contribution | -27.88 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.16 |
| Mean z-score | -4.34 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.62 |
| SVM RNA-class probability | 0.999055 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15716534 120 - 22422827 AUACACGGGUGCGAUAGAUCGAAAAUCCCUUCAUCAACAAAGUGUCCAAAAGUAUGCUAUAGAAUAUCUCGAAUAUGCCAUAUUCUUCGUUGCGUACUUAUGGACACCCAAAAGCCUCCG .....((((.((....(((.(((......))))))......(((((((.((((((((...(((((((..(......)..))))))).....)))))))).)))))))......)).)))) ( -35.80, z-score = -5.11, R) >droEre2.scaffold_4690 7422480 117 + 18748788 AUACAUGGGUGCGAUAGCUCGAAAAUCCC---UUCAACAGACUGUCUGAAAGUAUGCUAUAGAAUAUCUUGACCAUGCCAUGUUCUUCACUGCAUACUUUCGGACACCUUACAGCCUCCG ......(((.((........(((......---))).......(((((((((((((((...(((((((............))))))).....))))))))))))))).......)).))). ( -32.90, z-score = -3.59, R) >droSec1.super_37 297484 117 - 454039 AUACACGGGUGCGAUAGAUCGAAAAUCCC---UUCAACAAAGUGUCCAAAAGUAUGCUACAGAAUAUGGCGGCUAUGCCAUAUUCUUCGUUGCGUACUCACGGACACCCAAAAGCCUACG ......(((..(((....))).....)))---.........((((((...(((((((.(((((((((((((....)))))))))))..)).)))))))...))))))............. ( -40.20, z-score = -4.98, R) >droSim1.chrU 78462 116 - 15797150 AUACACGGGUGCGAUAGAUCGAAAAUCCC----UCAACAAAGUGUCCAAAAGUAUGCUACAGAAUAUCUCGGCUAUGCCAUAUUAUUCGUUGCGUACUUACGGACACCCAAAAGCCUGCG .....(((((..((..(((.....)))..----))......((((((..((((((((.((.(((((....(((...)))....))))))).))))))))..))))))......))))).. ( -34.60, z-score = -3.67, R) >consensus AUACACGGGUGCGAUAGAUCGAAAAUCCC___UUCAACAAAGUGUCCAAAAGUAUGCUACAGAAUAUCUCGACUAUGCCAUAUUCUUCGUUGCGUACUUACGGACACCCAAAAGCCUCCG ......((((.(((....)))...)))).............(((((((.((((((((...(((((((..((....))..))))))).....)))))))).)))))))............. (-27.50 = -27.88 + 0.38)

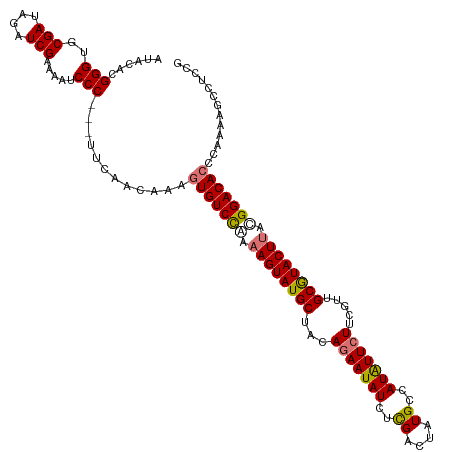
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:47:40 2011