| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,476,094 – 11,476,192 |
| Length | 98 |
| Max. P | 0.962897 |

| Location | 11,476,094 – 11,476,192 |
|---|---|
| Length | 98 |
| Sequences | 9 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 76.38 |
| Shannon entropy | 0.49072 |
| G+C content | 0.56090 |
| Mean single sequence MFE | -27.47 |
| Consensus MFE | -18.46 |
| Energy contribution | -18.97 |
| Covariance contribution | 0.51 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.962897 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 11476094 98 + 23011544 ---AAAUGGACACGCACACGUGGUCAAUUGCGUU-GGGGUCAACCCUUUCGCACCAGCGCCAUCAUUCUGCUGUCCAGCCUUCUCUCCACCUCCAGCUCCAG ---...((((((.(((...(((((....((((..-((((....))))..)))).....))))).....)))))))))......................... ( -29.40, z-score = -2.51, R) >droSim1.chr2L 11288379 98 + 22036055 ---AAAUGGACACGCACACGUGGUCAAUUGCGUU-GGGGUCAACCCUUUCGCACCAGCGCCAUCAUUCUGCUGUCCAGCCUUCUGCCCACCUCCAGCUCCAG ---...((((((.(((...(((((....((((..-((((....))))..)))).....))))).....)))))))))......................... ( -29.40, z-score = -1.67, R) >droSec1.super_3 6871282 98 + 7220098 ---AAAUGGACACGCACACGUGGUCAAUUGCGUU-GGGGUCAACCCUUUCGCAUCAGCGCCAUCAUUCUGCUGUCCAGCCUUCUGCCCACCUCCAUCUCCAG ---...((((((.(((...(((((....((((..-((((....))))..)))).....))))).....)))))))))......................... ( -30.10, z-score = -2.71, R) >droEre2.scaffold_4929 12683545 98 - 26641161 ---AAAUGGACACGCACACGUGGUCAAUUGCGUU-GGGGUCAACCCUUUCGCACCAGCGCCAUCAUUCUGCUGUCCAGCCUUCCCUCCAGCUCCAGCUCCAG ---...((((((.(((...(((((....((((..-((((....))))..)))).....))))).....)))))))))...........(((....))).... ( -30.90, z-score = -2.43, R) >droAna3.scaffold_12916 12179381 95 - 16180835 ---AAAUGGACACGCACACGUGGUCAAUUGCGUUUGGGGUCAACCCUUUCGCACCAGCGCCAUCAUUCUGCUGUCCAGUGUCCCAUUC--CCCCAUUUUC-- ---...((((((.(((...(((((....((((...((((....))))..)))).....))))).....)))))))))...........--..........-- ( -28.10, z-score = -2.20, R) >droWil1.scaffold_181038 374230 95 - 637489 ---AAGUGGACACGCACACGUGGUCAAUU--GUUUGGGGUCAACCCUUUCGCACCAAUGCCAUUACUCUACACCCCCACA-CGACACCAACACCGUCUCCA- ---...((((.(((......((((...((--((.(((((...........(((....))).............))))).)-))).))))....))).))))- ( -21.55, z-score = -0.56, R) >droVir3.scaffold_12963 18142345 85 - 20206255 ---AAAUGGAAAAGCACACGUGGUCAAUUGCGUUUGGGGUCAACCCUUUCGCACGAACGCCAUCAUUCCGU--GCCGGCUCCUGCCCCAC------------ ---...(((....((((..(((((....((((...((((....))))..)))).....)))))......))--)).(((....)))))).------------ ( -26.70, z-score = -1.00, R) >droMoj3.scaffold_6500 5147106 80 - 32352404 ---AAACGGUAAAGCACACGUGGUCAAUUGCGUUUGGGGUCAACCCUUUCGCACGAACGCCAUCAUUCUGC--ACCGGCCACUGC----------------- ---...((((...(((...(((((....((((...((((....))))..)))).....))))).....)))--))))........----------------- ( -23.10, z-score = -0.57, R) >droGri2.scaffold_15126 7314050 94 - 8399593 AAGAAAUGGCAGAGAACACGUGGUCAAUUGCGUUUGGGGUCAACCCUUUCGCACGAACGCCAUCAUUCUGC--ACCGGCGCCUGCCCCACACCACA------ .......(((((((((...(((((....((((...((((....))))..)))).....)))))..))))((--....))..)))))..........------ ( -28.00, z-score = -0.54, R) >consensus ___AAAUGGACACGCACACGUGGUCAAUUGCGUUUGGGGUCAACCCUUUCGCACCAGCGCCAUCAUUCUGCUGUCCAGCCUCCGCCCCACCUCCAGCUCCA_ ......((((...(((...(((((....((((...((((....))))..)))).....))))).....)))..))))......................... (-18.46 = -18.97 + 0.51)

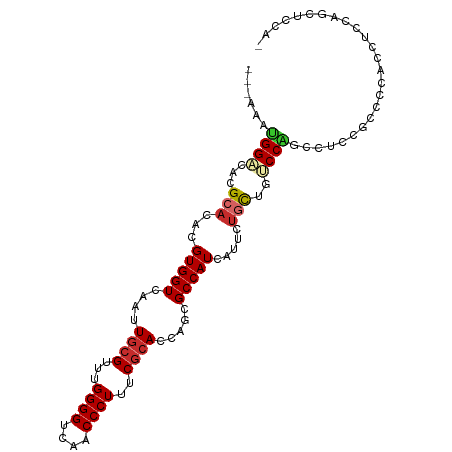
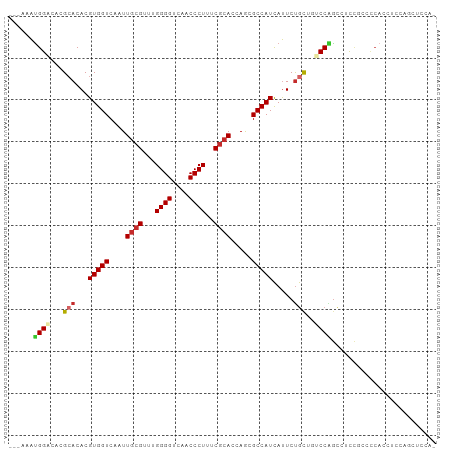
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:32:44 2011