| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,713,113 – 15,713,284 |
| Length | 171 |
| Max. P | 0.530005 |

| Location | 15,713,113 – 15,713,284 |
|---|---|
| Length | 171 |
| Sequences | 3 |
| Columns | 196 |
| Reading direction | forward |
| Mean pairwise identity | 70.91 |
| Shannon entropy | 0.40966 |
| G+C content | 0.39828 |
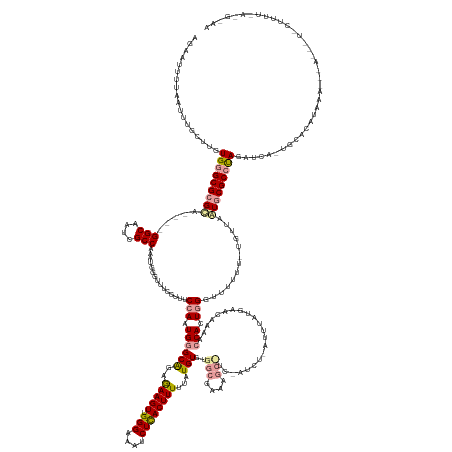
| Mean single sequence MFE | -50.77 |
| Consensus MFE | -27.66 |
| Energy contribution | -28.37 |
| Covariance contribution | 0.71 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.530005 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
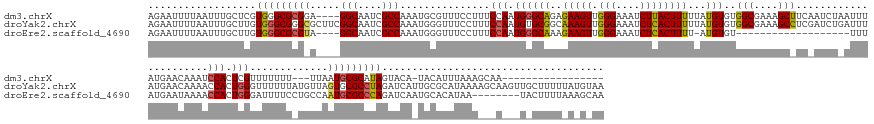
>dm3.chrX 15713113 171 + 22422827 AGAAUUUUAAUUUGCUCGUGGGCGCGGA----GGCAAUCGCCAAAUGCGUUUCCUUUCCAAUGGGCAGAGAAGUUGGGAAAUCUUACUUUUUAUGUGUGGCGAAAGCUUCAAUCUAAUUUAUGAACAAAUCCACUCGUUUUUUU---UUAAUGCGCAUAGUACA-UACAUUUAAAGCAA----------------- ..((((((..((((((((((((((((..----(((....)))...))))).......)).)))))))))))))))...........((((..(((((((((....))...........(((((..((.................---....))..)))))..))-)))))..))))...----------------- ( -38.21, z-score = 0.32, R) >droYak2.chrX 9912010 196 + 21770863 AGAAUUUUAAUUUGCUUGUGGGCGGGCGCUUCGGCAAUCGCCAAAUGGGUUUCCUUUCCAAUGUGCGGCAAAGUUGGGAAAUCUCACUUUUUAUGUGUGGCGAAAGCCUCGAUCUGAUUUAUGAACAAAACCACUGGGUUUUUUAUGUUAGUGCGCCUAGAUCAUUGCGCAUAAAAGCAAGUUGCUUUUUAUGUAA .......((((((((((((((((((..((....))..)))))....(((((((((..(...((.....))..)..))))))))))))...(((((((((((....)))..((((((...(((.(((((((((....)))))....)))).)))....))))))...))))))))))))))))))............ ( -62.20, z-score = -2.15, R) >droEre2.scaffold_4690 7419306 164 - 18748788 AGAAUUUUAAUUUGCUUGUGGGCGCGUA----GGCAAUCGCCAAAUGGGUUUCCUUUCCAAUGGGCAAAGAAGUUGGGAAAUCUCACUUUU-AUGUGU-------------------UUUAUGAAUAAAACCACUGGGAUUUUCCUGCCAAUGCGCCCAGAUCAAUGCACAUAA--------UACUUUUAAAGCAA ...........((((((.(((((((((.----((((...(((...((((.......))))...))).........(((((((((((.....-.((.((-------------------((((....)))))))).))))))))))))))).)))))))))............(((--------.....))))))))) ( -51.90, z-score = -2.95, R) >consensus AGAAUUUUAAUUUGCUUGUGGGCGCGCA____GGCAAUCGCCAAAUGGGUUUCCUUUCCAAUGGGCAGAGAAGUUGGGAAAUCUCACUUUUUAUGUGUGGCGAAAGC_UC_AUCU_AUUUAUGAACAAAACCACUGGGUUUUUU_UGUUAAUGCGCCUAGAUCA_UGCACAUAAA___A___U_CUUUU_A_G_AA ..................(((((((((.....(((....)))...............(((.((((((..(((((.(((....))))))))...)))..(((....)))......................))).))).............)))))))))..................................... (-27.66 = -28.37 + 0.71)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:47:36 2011