
| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,712,443 – 15,712,570 |
| Length | 127 |
| Max. P | 0.794554 |

| Location | 15,712,443 – 15,712,544 |
|---|---|
| Length | 101 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 69.29 |
| Shannon entropy | 0.60383 |
| G+C content | 0.39770 |
| Mean single sequence MFE | -24.67 |
| Consensus MFE | -9.89 |
| Energy contribution | -9.16 |
| Covariance contribution | -0.73 |
| Combinations/Pair | 1.58 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.794554 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15712443 101 - 22422827 CUCCUUGAAUUCCUCGAGGGAAUCUAAAUGAUUUGAGAAGGGCGCCACUGUAAAGCCGAUAUCGC-UGGUGAUAUUCCAAAAUAAGAUUCAAAA-AACUCUUA--------------- ..((((((.....))))))((((((...((.((((.(((...(((((.((............)).-)))))...))))))).))))))))....-........--------------- ( -22.50, z-score = -0.91, R) >droSim1.chrX_random 4072230 102 - 5698898 CUCCUUGAAUUCCUCGAGGGAAUCUAAAUGAUUUGAGAAGGGCGCCACUGUAAAGCCGAUAUCGCUUGCUGAUAUUCCAAAAUAAGAUUCAAAA-AACGCUAA--------------- ..((((((.....))))))((((((...((.((((.(((.(((...........)))..(((((.....)))))))))))).))))))))....-........--------------- ( -20.40, z-score = -0.29, R) >droSec1.super_37 293517 102 - 454039 CUCCUUGAAUUCCUCGAGGGAAUCUAAAUGAUUUGAGAAGGGCGCCACUGUAAAGCCGAUAUCGCUUGGUGAUAUUCCAAAAUAAGCUUCAAAA-AACGCCUA--------------- ..((((((.....))))))(((((.....)))))..(((((((...........)))((((((((...))))))))..........))))....-........--------------- ( -23.00, z-score = -0.62, R) >droYak2.chrX 9911328 100 - 21770863 UUCCUUGAAUUCCUCGAGGGAAUCUAAGUGAUUUGAGAAUGGCGCCACUGUAAAGCCGAUAUCGC-UGAUGAUACACC-AAAUAAGAUUCAAAA-AACUCGUA--------------- ..((((((.....))))))((((((.....(((((....((((...........)))).(((((.-...)))))...)-)))).))))))....-........--------------- ( -20.20, z-score = -0.71, R) >droEre2.scaffold_4690 7418635 100 + 18748788 CUCCUUGAAUUCCUCGAGGGAAUCUAAGUGAUUUGAGAAAGGCGCCACUGUAAAGCCGAUAUCGC-UGAUGAUACUCC-AAAUAAGAUUCAAAA-AACUCUUA--------------- ..((((((.....))))))((((((.....(((((((...(((...........)))..(((((.-...))))))).)-)))).))))))....-........--------------- ( -21.00, z-score = -0.99, R) >droAna3.scaffold_13417 4180731 114 + 6960332 UGCCUUAAAUUCCACAAGGGAAUCCAAAUGGUUAGAGAACGGCGCCACUUUAUAGCUGUUUUUU--UAAU-ACUUCCA-GACAAACUACUAGUAGAAAGAACAAAUUAAGUUGGUAUU .(((....(((((.....)))))((....)).........)))((((.((((....((((((((--(..(-(((...(-(.....))...)))))))))))))...)))).))))... ( -21.90, z-score = -0.66, R) >dp4.Unknown_group_104 5303 96 + 31585 UGCCUUGAAUUCCUCAAGGGAAUCCAAGUGGUUAGAGAACGGCGCCAACUUAAAGCCGUUUUCU--UAAU---UUAAA-AAAAAACUUGUAAUUAGAAGUUA---------------- ...((((.(((((.....))))).)))).((((((((((((((...........))))))))).--))))---)....-....(((((.(....).))))).---------------- ( -25.00, z-score = -2.62, R) >droPer1.super_95 117200 95 - 176753 UGCCUUGAAUUCCUCAAGGGAAUCCAAGUGGUUAGAGAACGGCGCCAACUUAAAGCCGUUUUCU--UAAU---UU-AA-AAAAAACUUGUAAUUAGAAGUUA---------------- ...((((.(((((.....))))).)))).((((((((((((((...........))))))))).--))))---).-..-....(((((.(....).))))).---------------- ( -25.00, z-score = -2.58, R) >droWil1.scaffold_181096 6014852 95 - 12416693 UGUUUUGAAUUCCUCAAGGGAAUCCAAAUGCUUAGAGAACGGCGCCACCUUAGAGCCGUUGUCG--AAAUCGUUUUUU-AAAUAAAUUUUGAUAGAAU-------------------- .((((((.(((((.....))))).)))).))...((.((((((...........)))))).)).--.....(((((((-(((.....))))).)))))-------------------- ( -20.40, z-score = -1.00, R) >droVir3.scaffold_12970 11655311 99 - 11907090 AGCCUUGAAUUCCUCGAGGGAAUCCAAAUGUUUGGAGAACGGCGCCACUUUGGAGCCGUUGUCUA-AGUCGCUUUUC--AAAUAAAUUUUGGAA-AAUUCACA--------------- ..((((((.....))))))((((......(((((((.((((((.((.....)).)))))).))))-)).)..(((((--(((.....)))))))-)))))...--------------- ( -32.30, z-score = -2.85, R) >droMoj3.scaffold_6473 7356211 88 + 16943266 GGUCUUAAAUUCCUCGAGGGAAUCCAAGUGCUUGGAGAACGGCGCCAAUUUGGAGCCGUUAUCU--GAGUCGCUCUUC-AGAUAAAUCUUG--------------------------- .((((...(((((.....)))))...((((((..((.((((((.((.....)).)))))).)).--.)).))))....-))))........--------------------------- ( -29.20, z-score = -2.25, R) >droGri2.scaffold_15203 3627515 100 - 11997470 AGCCUUGAAUUCCUCGAGGGAAUCCAAAUGUUUGGAGAACGGCGCCACUUUGGAGCCGUUGUCUG-UGUCGUUUUUC--AAAUAAAUUUUGGAAUAAUUCUCA--------------- ..((((((.....))))))...(((((((((((((((((((((((.((..((....))..))..)-)))))))))))--)))))...))))))..........--------------- ( -35.10, z-score = -3.70, R) >consensus UGCCUUGAAUUCCUCGAGGGAAUCCAAAUGAUUUGAGAACGGCGCCACUUUAAAGCCGUUAUCG__UGAUGAUUUUCC_AAAUAAGUUUUAAAA_AACGCUUA_______________ ..((((((.....))))))..............(((....(((...........)))....)))...................................................... ( -9.89 = -9.16 + -0.73)

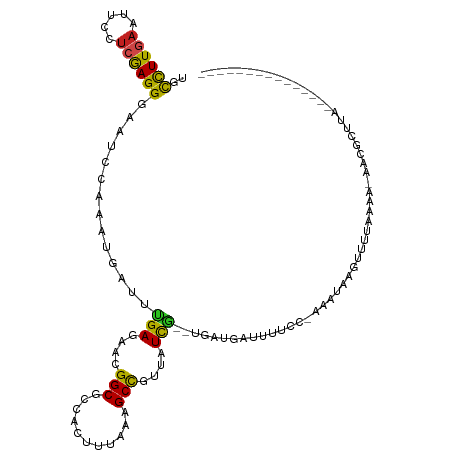

| Location | 15,712,458 – 15,712,570 |
|---|---|
| Length | 112 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 71.60 |
| Shannon entropy | 0.59366 |
| G+C content | 0.43627 |
| Mean single sequence MFE | -25.38 |
| Consensus MFE | -11.64 |
| Energy contribution | -11.16 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.64 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.602215 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15712458 112 - 22422827 --CAGAUGCCAAUCCACUCACCCGAUAACUCCUUGAAUUCCUCGAGGGAAUCUAAAUGAUUUGAGAAGGGCGCCACUGUAAAGCCGAUAUCGC-UGGUGAUAUUCCAAAAUAAGA-- --..(((....)))...((((((((((...((((((.....))))))(((((.....)))))......(((...........)))..))))).-.)))))...............-- ( -24.00, z-score = -0.13, R) >droSim1.chrX_random 4072245 113 - 5698898 --CAGAUGCCAAUCCACUCACCGGAUAACUCCUUGAAUUCCUCGAGGGAAUCUAAAUGAUUUGAGAAGGGCGCCACUGUAAAGCCGAUAUCGCUUGCUGAUAUUCCAAAAUAAGA-- --(((..((..(((..(((...........((((((.....))))))(((((.....))))))))...(((...........))))))...))...)))................-- ( -21.90, z-score = 0.61, R) >droSec1.super_37 293532 113 - 454039 --CAGAUGCCAAUCCACUCACCCGAUAACUCCUUGAAUUCCUCGAGGGAAUCUAAAUGAUUUGAGAAGGGCGCCACUGUAAAGCCGAUAUCGCUUGGUGAUAUUCCAAAAUAAGC-- --..(((....)))...((((((((((...((((((.....))))))(((((.....)))))......(((...........)))..)))))...)))))...............-- ( -23.60, z-score = 0.02, R) >droYak2.chrX 9911343 111 - 21770863 --UAGAUGCCAAUCCACUCACCCGAUAAUUCCUUGAAUUCCUCGAGGGAAUCUAAGUGAUUUGAGAAUGGCGCCACUGUAAAGCCGAUAUCGC-UGAUGAUACACC-AAAUAAGA-- --..(.(((((.((((.((((..(((..((((((((.....)))))))))))...))))..)).)).))))).)..((((.(((.(....)))-).....))))..-........-- ( -23.90, z-score = -1.14, R) >droEre2.scaffold_4690 7418650 111 + 18748788 --CAGAUGCCAAUUCACUCACCCGAUAACUCCUUGAAUUCCUCGAGGGAAUCUAAGUGAUUUGAGAAAGGCGCCACUGUAAAGCCGAUAUCGC-UGAUGAUACUCC-AAAUAAGA-- --((...(((((.(((((.....(((...(((((((.....))))))).)))..))))).))).....(((...........)))......))-...)).......-........-- ( -20.30, z-score = 0.31, R) >droAna3.scaffold_13417 4180763 104 + 6960332 ------UGCCAGCUAACUUACCAGAAAGUGCCUUAAAUUCCACAAGGGAAUCCAAAUGGUUAGAGAACGGCGCCACUUUAUAGCUGUUUUUU--UAAUA--CUUCC-AGACAAAC-- ------...((((((.((....))...(((((....(((((.....)))))((....)).........))))).......))))))......--.....--.....-........-- ( -18.20, z-score = 0.07, R) >dp4.Unknown_group_104 5318 94 + 31585 ---------------ACUCACCAGAUAAUGCCUUGAAUUCCUCAAGGGAAUCCAAGUGGUUAGAGAACGGCGCCAACUUAAAGCCGUUUUCU--UAA-----UUUA-AAAAAAAACU ---------------..........((((..((((.(((((.....))))).))))..))))(((((((((...........))))))))).--...-----....-.......... ( -24.80, z-score = -3.10, R) >droPer1.super_95 117215 108 - 176753 UGCCGAUAGCACCGAACUCACCAGAUAAUGCCUUGAAUUCCUCAAGGGAAUCCAAGUGGUUAGAGAACGGCGCCAACUUAAAGCCGUUUUCU--UAA-----UUUA-AAAAAAACU- (((.....)))...(((.(((..(((..(.((((((.....)))))).))))...))))))((((((((((...........))))))))))--...-----....-.........- ( -26.50, z-score = -2.26, R) >droWil1.scaffold_181096 6014862 108 - 12416693 -----GUAAAAGUAAACUCACCCGAUAGUGUUUUGAAUUCCUCAAGGGAAUCCAAAUGCUUAGAGAACGGCGCCACCUUAGAGCCGUUGUCG--AAAUCGUUUUUU-AAAUAAAUU- -----.................(((((((((((.((.((((....)))).)).)))))))..((.((((((...........)))))).)).--..))))......-.........- ( -24.30, z-score = -1.79, R) >droVir3.scaffold_12970 11655325 100 - 11907090 -------------CAACUCACCGGUGAGAGCCUUGAAUUCCUCGAGGGAAUCCAAAUGUUUGGAGAACGGCGCCACUUUGGAGCCGUUGUCU--AAGUCGCUUUUC-AAAUAAAUU- -------------...((((....))))(((.(((.(((((.....))))).)))..(((((((.((((((.((.....)).)))))).)))--))).))))....-.........- ( -31.90, z-score = -2.12, R) >droMoj3.scaffold_6473 7356214 99 + 16943266 --------------CACUCACCGGACAGGGUCUUAAAUUCCUCGAGGGAAUCCAAGUGCUUGGAGAACGGCGCCAAUUUGGAGCCGUUAUCU--GAGUCGCUCUUC-AGAUAAAUC- --------------......((.....))((((...(((((.....)))))...((((((..((.((((((.((.....)).)))))).)).--.)).))))....-)))).....- ( -30.90, z-score = -1.32, R) >droGri2.scaffold_15203 3627530 108 - 11997470 -----CAAGUGUCAAACUCACCGGAUAGAGCCUUGAAUUCCUCGAGGGAAUCCAAAUGUUUGGAGAACGGCGCCACUUUGGAGCCGUUGUCU--GUGUCGUUUUUC-AAAUAAAUU- -----...(((.......))).((((....((((((.....))))))..))))...(((((((((((((((((.((..((....))..))..--))))))))))))-)))))....- ( -34.20, z-score = -2.37, R) >consensus _____AUGCCAAUCCACUCACCCGAUAAUGCCUUGAAUUCCUCGAGGGAAUCCAAAUGAUUUGAGAACGGCGCCACUUUAAAGCCGUUAUCG__UAAUGAUAUUCC_AAAUAAAA__ ......................(((((...((((((.....)))))).....................(((...........)))..)))))......................... (-11.64 = -11.16 + -0.48)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:47:34 2011