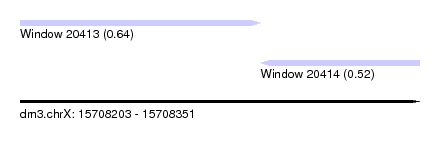
| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,708,203 – 15,708,351 |
| Length | 148 |
| Max. P | 0.638151 |

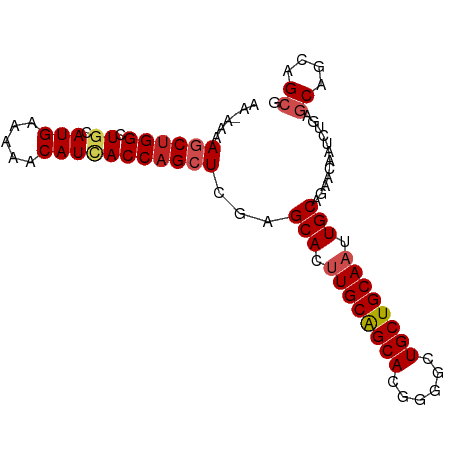
| Location | 15,708,203 – 15,708,292 |
|---|---|
| Length | 89 |
| Sequences | 11 |
| Columns | 89 |
| Reading direction | forward |
| Mean pairwise identity | 89.58 |
| Shannon entropy | 0.21559 |
| G+C content | 0.52227 |
| Mean single sequence MFE | -28.67 |
| Consensus MFE | -24.23 |
| Energy contribution | -24.78 |
| Covariance contribution | 0.55 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.638151 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
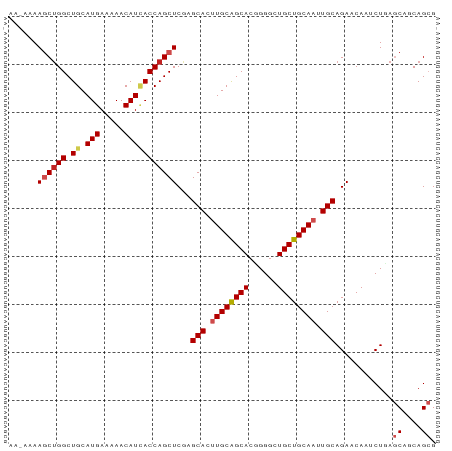
>dm3.chrX 15708203 89 + 22422827 AAAAAAAGCUGGCUGCAUGAAAAACAUCACCAGCUCGAGCACUUGCAGCACUGGGCUGCUGCAAUUGCAGAACAACCUGAGCUGCAGCG ...........(((((.(((......)))..(((((..(((.((((((((......)))))))).)))((......)))))))))))). ( -32.40, z-score = -1.96, R) >droEre2.scaffold_4690 7414369 89 - 18748788 AAAAAAAGCUGGCUGCAUGAAAAACAUCACCAGCUCGAGCACUUGCAGCACCGGGCUGCUGCAAUUGCAGAACAACCUGAGCUGCAGCG ...........(((((.(((......)))..(((((..(((.((((((((......)))))))).)))((......)))))))))))). ( -32.40, z-score = -2.10, R) >droYak2.chrX 9907202 89 + 21770863 AGAAAAAGCUGGCUGCAUGAAAAACAUCACCAGCUCGAGCACUUGCAGCACCGGGCUGCUGCAAUUGCAGAACAACCUGAGCUGCAGCG ...........(((((.(((......)))..(((((..(((.((((((((......)))))))).)))((......)))))))))))). ( -32.40, z-score = -1.74, R) >droSec1.super_37 289379 89 + 454039 AAGAAAAGCUGGCUGCAUGAAAAACAUCACCAGCUCGAGCACUUGCAGCACCGGGCUGCUGCAAUUGCAGAACAACCUGAGCUGCAGCG ...........(((((.(((......)))..(((((..(((.((((((((......)))))))).)))((......)))))))))))). ( -32.40, z-score = -1.74, R) >droSim1.chrX_random 4067191 89 + 5698898 AAGAAAAGCUGGCUGCAUGAAAAACAUCACCAGCUCGAGCACUUGCAGCACCGGGCUGCUGCAAUUGCAGAACAACCUGAGCUGCAGCG ...........(((((.(((......)))..(((((..(((.((((((((......)))))))).)))((......)))))))))))). ( -32.40, z-score = -1.74, R) >droMoj3.scaffold_6473 7342772 89 - 16943266 GUAAAAAGCUGGCUGCAUGAAAAACAUCACCAGCUCAAGCACUUGCGGCACGGGGCUGCUGCAAUUGCAGAACAAUCUCAGCAACAGCG ......((((((.((.(((.....)))))))))))...(((.((((((((......)))))))).)))............((....)). ( -27.80, z-score = -1.27, R) >droVir3.scaffold_12970 11642609 89 + 11907090 AAUAAAAGCUGGCUGCAUGAAAAACAUCACCAGCUCAAGCACUUGCGGCACGGGGCUGCUGCAAUUGCAGAACAAUCUCAGCAACAGCG ......((((((.((.(((.....)))))))))))...(((.((((((((......)))))))).)))............((....)). ( -27.80, z-score = -1.56, R) >droGri2.scaffold_15203 3615191 84 + 11997470 -----AAACUGGCUGCAUGAAAAACAUCACCAGCUCAAGCACUUGCGGCACGGGGCUGCUGCAAUUGCAGAACAAUCUCAGCAACAGCG -----...((((((((((((......)))...((....))...)))))).))).((((.(((...((.(((....)))))))).)))). ( -23.90, z-score = -0.76, R) >droWil1.scaffold_181096 6000290 84 + 12416693 -----AAACGGGCUGCAUGAAAAACAUUACCAGCUCAAGCACUUGCAGCACUGGCCUGCUGCAAUUGCAGAACAAUCUCAGCAGCAGCG -----......((((((((.....))).....(((...(((.((((((((......)))))))).))).((......))))).))))). ( -27.30, z-score = -1.38, R) >droPer1.super_95 100261 84 + 176753 -----AAGCUGGCUGCAUGAAAAACAUAACCAGCUCGAGCACCUGCAGCACGGGGCUGCUGCAAUUGCAGAACAAUCUAAACAACAGGG -----..((((((((.(((.....)))...)))))..))).((((((((.....))))((((....))))..............)))). ( -23.30, z-score = -0.72, R) >dp4.Unknown_group_9 52432 84 + 53872 -----AAGCUGGCUGCAUGAAAAACAUAACCAGCUCGAGCACCUGCAGCACGGGGCUGCUGCAAUUGCAGAACAAUCUAAACAACAGGG -----..((((((((.(((.....)))...)))))..))).((((((((.....))))((((....))))..............)))). ( -23.30, z-score = -0.72, R) >consensus AA_AAAAGCUGGCUGCAUGAAAAACAUCACCAGCUCGAGCACUUGCAGCACGGGGCUGCUGCAAUUGCAGAACAAUCUGAGCAGCAGCG ......((((((.((.(((.....)))))))))))...(((.((((((((......)))))))).)))............((....)). (-24.23 = -24.78 + 0.55)

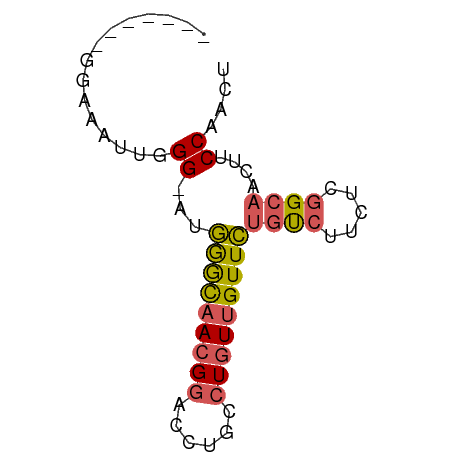
| Location | 15,708,292 – 15,708,351 |
|---|---|
| Length | 59 |
| Sequences | 5 |
| Columns | 67 |
| Reading direction | reverse |
| Mean pairwise identity | 71.60 |
| Shannon entropy | 0.50493 |
| G+C content | 0.51377 |
| Mean single sequence MFE | -17.74 |
| Consensus MFE | -9.54 |
| Energy contribution | -9.78 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.519210 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15708292 59 - 22422827 -------GGUAAUUCGG-AUGGGCAACGGACCUGCCUGUUGUUCUGUCUUCUCGGCAACUUCCAACU -------........((-(.(((((((((......)))))))))((((.....))))...))).... ( -16.20, z-score = -1.20, R) >droSec1.super_37 289468 59 - 454039 -------GGUGAUUUGG-AUGGGCAACGGACCUGCCUGUUGUUCUGUCUUCUCGGCAACUUCCAACU -------......((((-(.(((((((((......)))))))))((((.....))))...))))).. ( -17.40, z-score = -1.32, R) >droYak2.chrX 9907291 67 - 21770863 AAUGAGGGGGAAUGGGGAAUGGGCAACGGACCUGCCUGUUGUUCUGUCUUCUCGGCAACUUCCAACU ....((..((((.(((((..(((((((((......)))))))))..)))))..(....)))))..)) ( -21.80, z-score = -0.95, R) >droEre2.scaffold_4690 7414458 65 + 18748788 GGUGAUUGGAAA--GGGAAUGGGCAACGGACCUGCCUGUUGUUCUGUCUUCUCGGCAACUUCCAACU .....((((((.--(((((.(((((((((......)))))))))....)))))(....))))))).. ( -21.30, z-score = -1.53, R) >droMoj3.scaffold_6473 7342861 59 + 16943266 --------GAGAAAUGGUGUAAAUAACGCACCUGCUUCUGGUUUGCCAACAACAGCGACGUCCAAUU --------.((((..(((((.......)))))...))))((.((((........))))...)).... ( -12.00, z-score = -0.47, R) >consensus _______GGAAAUUGGG_AUGGGCAACGGACCUGCCUGUUGUUCUGUCUUCUCGGCAACUUCCAACU ...............((...(((((((((......)))))))))((((.....))))....)).... ( -9.54 = -9.78 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:47:32 2011