
| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,698,221 – 15,698,382 |
| Length | 161 |
| Max. P | 0.949075 |

| Location | 15,698,221 – 15,698,382 |
|---|---|
| Length | 161 |
| Sequences | 5 |
| Columns | 171 |
| Reading direction | forward |
| Mean pairwise identity | 76.46 |
| Shannon entropy | 0.39855 |
| G+C content | 0.40036 |
| Mean single sequence MFE | -40.08 |
| Consensus MFE | -19.44 |
| Energy contribution | -22.24 |
| Covariance contribution | 2.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.923489 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
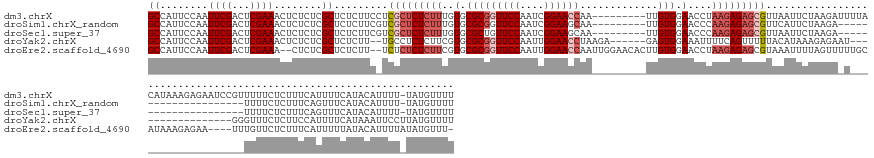
>dm3.chrX 15698221 161 + 22422827 AAAACAUA-AAAAUGUAUGAAAAUGAAAGAGAAAAACGGAUUCUCUUUAUGUAAAAUCUUAGAAUUAACGCUCUCUUAGGUUCCACAA---------UUGGUUCCGAUUGGAACCGCGCACAAAGAGAGCGAGGAAGAGAGCGAGAGAGUUUCGAGUCGAAUUGGAAUGGC ...(((((-((....(((....)))...(((((.......))))))))))))................(((((((((.(((((((..(---------(((....))))))))))).(((.(.....).)))...))))))))).....(((((((......)))))))... ( -43.70, z-score = -2.67, R) >droSim1.chrX_random 4478 140 + 5698898 AAAACAUA-AAAAUGUAUGAAACUGAAAGAGAAAA---------------------UCUUAGAAUGAACGCUCUCUUGGGUUCCACAA---------UUGCUUCCGAUUGGAACCGCGCACAAAGAGAGCGACGAAGAGAGCGAGAGAGUUUCGAGUCGAAUUGGAAUGGC ...((((.-...)))).(((((((..((((.....---------------------))))........(((((((((.(((((((..(---------(((....)))))))))))((((.(.....).))).).)))))))))....)))))))................. ( -38.00, z-score = -2.19, R) >droSec1.super_37 278823 140 + 454039 AAAACAUA-AAAAUGUAUGAAACUGAAAGAGAAAA---------------------UCUUAGAAUUAACGCUCUCUUGGGUUCCACAA---------UUGCUUCCGAUUGGAACAGCGCACAAAGAGAGCGACGAAGAGAGCGAGAGAGUUUCGAGUCGAAUUGGAAUGGC ...((((.-...)))).(((((((..((((.....---------------------))))........(((((((((.(((((((..(---------(((....))))))))))..(((.(.....).))).).)))))))))....)))))))................. ( -36.60, z-score = -2.00, R) >droYak2.chrX 9897131 146 + 21770863 AAAACAUAAGGAAUUUAUGAAAAUGGAAGAGAAACCC-----------------AUUCUCUUUAUGUAAAAACUGAAAAUUUCCACUC------UCUUAGGUUCCAAUUGGAACCGCGCACGAAGAGAGGCA--AAGAGAGCGAGAGAGUUUCGAGUCGAAUUGGAAUGGC ...((((((....(((((....)))))((((((....-----------------.))))))))))))....(((((((.(((((.(((------((((.((((((....))))))..((.(.......))).--))))))).).)))).)))).))).............. ( -38.80, z-score = -2.23, R) >droEre2.scaffold_4690 7404421 162 - 18748788 -AAACAUAUAAAAUGUAUAAAAAUGAAAGAGAACAAA----UUCUCUUUAUGCAAAAACUAAAAUUUACGCUCUCUUAGGUUCCACAAGUGUUCCAAUUGGUUCCAAUUGGAACCGCGCACGAAGAGAGAGA--AAGAGAGCGAGAG--UUUCGAGUCGAAUUGGAAUGGC -...(((......(((((((.......((((((....----)))))))))))))...(((.((((((.(((((((((...(((.....((((((((((((....)))))))).....)))).....)))...--))))))))).)))--)))..))).........))).. ( -43.31, z-score = -3.28, R) >consensus AAAACAUA_AAAAUGUAUGAAAAUGAAAGAGAAAA___________________A_UCUUAGAAUUAACGCUCUCUUAGGUUCCACAA_________UUGGUUCCGAUUGGAACCGCGCACAAAGAGAGCGA_GAAGAGAGCGAGAGAGUUUCGAGUCGAAUUGGAAUGGC ....................................................................(((((((((......................((((((....)))))).(((.(.....).)))...))))))))).....(((((((......)))))))... (-19.44 = -22.24 + 2.80)



| Location | 15,698,221 – 15,698,382 |
|---|---|
| Length | 161 |
| Sequences | 5 |
| Columns | 171 |
| Reading direction | reverse |
| Mean pairwise identity | 76.46 |
| Shannon entropy | 0.39855 |
| G+C content | 0.40036 |
| Mean single sequence MFE | -36.34 |
| Consensus MFE | -16.20 |
| Energy contribution | -18.44 |
| Covariance contribution | 2.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.949075 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15698221 161 - 22422827 GCCAUUCCAAUUCGACUCGAAACUCUCUCGCUCUCUUCCUCGCUCUCUUUGUGCGCGGUUCCAAUCGGAACCAA---------UUGUGGAACCUAAGAGAGCGUUAAUUCUAAGAUUUUACAUAAAGAGAAUCCGUUUUUCUCUUUCAUUUUCAUACAUUUU-UAUGUUUU ..................((((......(((((((((((.(((.(.....).))).((((((....))))))..---------....))).....))))))))....................((((((((.......))))))))...))))..((((...-.))))... ( -36.80, z-score = -2.81, R) >droSim1.chrX_random 4478 140 - 5698898 GCCAUUCCAAUUCGACUCGAAACUCUCUCGCUCUCUUCGUCGCUCUCUUUGUGCGCGGUUCCAAUCGGAAGCAA---------UUGUGGAACCCAAGAGAGCGUUCAUUCUAAGA---------------------UUUUCUCUUUCAGUUUCAUACAUUUU-UAUGUUUU ..................((((((....(((((((((.(.(((.(.....).))))(((((((..(....)...---------...))))))).)))))))))........((((---------------------.....))))..))))))..((((...-.))))... ( -35.00, z-score = -2.69, R) >droSec1.super_37 278823 140 - 454039 GCCAUUCCAAUUCGACUCGAAACUCUCUCGCUCUCUUCGUCGCUCUCUUUGUGCGCUGUUCCAAUCGGAAGCAA---------UUGUGGAACCCAAGAGAGCGUUAAUUCUAAGA---------------------UUUUCUCUUUCAGUUUCAUACAUUUU-UAUGUUUU ..................((((((....(((((((((.(.(((.(.....).)))).((((((..(....)...---------...))))))..)))))))))........((((---------------------.....))))..))))))..((((...-.))))... ( -30.90, z-score = -1.85, R) >droYak2.chrX 9897131 146 - 21770863 GCCAUUCCAAUUCGACUCGAAACUCUCUCGCUCUCUU--UGCCUCUCUUCGUGCGCGGUUCCAAUUGGAACCUAAGA------GAGUGGAAAUUUUCAGUUUUUACAUAAAGAGAAU-----------------GGGUUUCUCUUCCAUUUUCAUAAAUUCCUUAUGUUUU .............((((.((((.(.((.(((((((((--.(((.(.....).).))((((((....)))))).))))------))))))).).))))))))..........((((((-----------------(((.......))))))))).................. ( -38.40, z-score = -3.26, R) >droEre2.scaffold_4690 7404421 162 + 18748788 GCCAUUCCAAUUCGACUCGAAA--CUCUCGCUCUCUU--UCUCUCUCUUCGUGCGCGGUUCCAAUUGGAACCAAUUGGAACACUUGUGGAACCUAAGAGAGCGUAAAUUUUAGUUUUUGCAUAAAGAGAA----UUUGUUCUCUUUCAUUUUUAUACAUUUUAUAUGUUU- .......(((...((((.((((--....(((((((((--....(((...((....))((((((((((....))))))))))......)))....)))))))))....)))))))).)))...((((((((----....)))))))).........((((.....))))..- ( -40.60, z-score = -3.28, R) >consensus GCCAUUCCAAUUCGACUCGAAACUCUCUCGCUCUCUUC_UCGCUCUCUUUGUGCGCGGUUCCAAUCGGAACCAA_________UUGUGGAACCUAAGAGAGCGUUAAUUCUAAGA_U___________________UUUUCUCUUUCAUUUUCAUACAUUUU_UAUGUUUU ..........((((...)))).......(((((((((...(((.(.....).))).((((((....))))))......................))))))))).................................................................... (-16.20 = -18.44 + 2.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:47:27 2011