
| Sequence ID | dm3.chrX |
|---|---|
| Location | 15,697,830 – 15,697,992 |
| Length | 162 |
| Max. P | 0.891518 |

| Location | 15,697,830 – 15,697,941 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 85.53 |
| Shannon entropy | 0.19028 |
| G+C content | 0.30396 |
| Mean single sequence MFE | -21.93 |
| Consensus MFE | -17.54 |
| Energy contribution | -17.77 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.764906 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15697830 111 + 22422827 UUAUAAACUAAUUUUAUACAUUGCUUAAGCGAUAGUUUGACGUAUAAAACUGAGGAGCUUAUAUUUAUUACUUUUGACUGCACAGUGGAAUGCAUUCAAAGGUCUGUUUAU ..((((((...(((((((((((((....))))).(.....)))))))))....................((((((((.((((........)))).))))))))..)))))) ( -22.40, z-score = -1.38, R) >droSim1.chrX_random 4103 96 + 5698898 UUAUAAACUCAUUUGAUACAUUGCUUAAGCGAUAGUUUGGUGU---------------CAAUAUUUAUUACUUUUGACUGCACAGUGGAAUGCAUUCAAGGGACUGUUUAU ..((((((....(((((((...(((........)))...))))---------------))).........(((((((.((((........)))).)))))))...)))))) ( -22.50, z-score = -2.07, R) >droSec1.super_37 278448 96 + 454039 UUAUAAACUCAUUUGAUACAUUGCUUAAGCGAUAGUUUGAUGU---------------UUAUAUUUAUUACUUUUGACUGCACAGUGGAAUGCAUUCAAAGGACUGUUUAU .((((((((((......(((((((....))))).)).))).))---------------))))).......(((((((.((((........)))).)))))))......... ( -20.90, z-score = -2.17, R) >consensus UUAUAAACUCAUUUGAUACAUUGCUUAAGCGAUAGUUUGAUGU_______________UUAUAUUUAUUACUUUUGACUGCACAGUGGAAUGCAUUCAAAGGACUGUUUAU ..((((((.((((....(((((((....))))).))..))))............................(((((((.((((........)))).)))))))...)))))) (-17.54 = -17.77 + 0.23)



| Location | 15,697,830 – 15,697,941 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 85.53 |
| Shannon entropy | 0.19028 |
| G+C content | 0.30396 |
| Mean single sequence MFE | -18.10 |
| Consensus MFE | -13.97 |
| Energy contribution | -13.97 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.847827 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15697830 111 - 22422827 AUAAACAGACCUUUGAAUGCAUUCCACUGUGCAGUCAAAAGUAAUAAAUAUAAGCUCCUCAGUUUUAUACGUCAAACUAUCGCUUAAGCAAUGUAUAAAAUUAGUUUAUAA ........((.(((((.(((((......))))).))))).))......((((((((....(((((((((((.....)....((....))...)))))))))))))))))). ( -23.50, z-score = -3.57, R) >droSim1.chrX_random 4103 96 - 5698898 AUAAACAGUCCCUUGAAUGCAUUCCACUGUGCAGUCAAAAGUAAUAAAUAUUG---------------ACACCAAACUAUCGCUUAAGCAAUGUAUCAAAUGAGUUUAUAA ((((((.(((..((((.(((((......))))).)))).((((.....)))))---------------))...........((....))..............)))))).. ( -15.10, z-score = -1.17, R) >droSec1.super_37 278448 96 - 454039 AUAAACAGUCCUUUGAAUGCAUUCCACUGUGCAGUCAAAAGUAAUAAAUAUAA---------------ACAUCAAACUAUCGCUUAAGCAAUGUAUCAAAUGAGUUUAUAA ...........(((((.(((((......))))).))))).........(((((---------------((...........((....))......((....))))))))). ( -15.70, z-score = -1.73, R) >consensus AUAAACAGUCCUUUGAAUGCAUUCCACUGUGCAGUCAAAAGUAAUAAAUAUAA_______________ACAUCAAACUAUCGCUUAAGCAAUGUAUCAAAUGAGUUUAUAA ((((((......((((.(((((......))))).))))...........................................((....))..............)))))).. (-13.97 = -13.97 + 0.00)

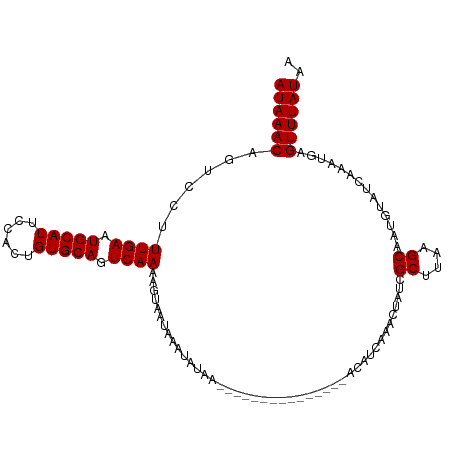

| Location | 15,697,868 – 15,697,980 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 83.80 |
| Shannon entropy | 0.21318 |
| G+C content | 0.40243 |
| Mean single sequence MFE | -24.50 |
| Consensus MFE | -23.29 |
| Energy contribution | -22.97 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.11 |
| Mean z-score | -0.81 |
| Structure conservation index | 0.95 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.589914 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15697868 112 + 22422827 GACGUAUAAAACUGAGGAGCUUAUAUUUAUUACUUUUGACUGCACAGUGGAAUGCAUUCAAAGGUCUGUUUAUGUUUUUGUUCCGGUGUGACCGUAUUCCGAACAGGAACAC ...............(((((..((((..((.((((((((.((((........)))).))))))))..))..))))....)))))((.....))((.((((.....)))).)) ( -26.60, z-score = -0.94, R) >droSim1.chrX_random 4141 97 + 5698898 ---------------GGUGUCAAUAUUUAUUACUUUUGACUGCACAGUGGAAUGCAUUCAAGGGACUGUUUAUGUUUUUGUUCCGGUGUGACCAUAUUCCGAACAGGGACAC ---------------.((((((((((..((..(((((((.((((........)))).)))))))...))..)))))(((((((.((.(((....))).)))))))))))))) ( -23.90, z-score = -0.62, R) >droSec1.super_37 278486 97 + 454039 ---------------GAUGUUUAUAUUUAUUACUUUUGACUGCACAGUGGAAUGCAUUCAAAGGACUGUUUAUGUUUUUGUUCCGGUGUGACCGUAAUCCGAACAGGGACGC ---------------.................(((((((.((((........)))).))))))).........(((((((((((((.....)))......)))))))))).. ( -23.00, z-score = -0.86, R) >consensus _______________GGUGUUUAUAUUUAUUACUUUUGACUGCACAGUGGAAUGCAUUCAAAGGACUGUUUAUGUUUUUGUUCCGGUGUGACCGUAUUCCGAACAGGGACAC ................................(((((((.((((........)))).)))))))........(((((((((((.((.(((....))).))))))))))))). (-23.29 = -22.97 + -0.33)

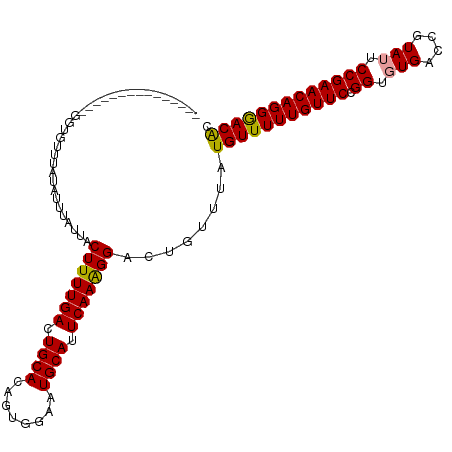
| Location | 15,697,880 – 15,697,992 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 89.63 |
| Shannon entropy | 0.13938 |
| G+C content | 0.38141 |
| Mean single sequence MFE | -28.00 |
| Consensus MFE | -25.29 |
| Energy contribution | -25.30 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.891518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15697880 112 + 22422827 UGAGGAGCUUAUAUUUAUUACUUUUGACUGCACAGUGGAAUGCAUUCAAAGGUCUGUUUAUGUUUUUGUUCCGGUGUGACCGUAUUCCGAACAGGAACACAGUCUAAUUUAA ...(((((..((((..((.((((((((.((((........)))).))))))))..))..))))....))))).....(((.((.((((.....)))).)).)))........ ( -30.10, z-score = -2.10, R) >droSim1.chrX_random 4146 104 + 5698898 --------CAAUAUUUAUUACUUUUGACUGCACAGUGGAAUGCAUUCAAGGGACUGUUUAUGUUUUUGUUCCGGUGUGACCAUAUUCCGAACAGGGACACAGUCUGAUUUAA --------..............(((((.((((........)))).)))))(((((((....((((((((((.((.(((....))).)))))))))))))))))))....... ( -26.60, z-score = -1.56, R) >droSec1.super_37 278491 104 + 454039 --------UUAUAUUUAUUACUUUUGACUGCACAGUGGAAUGCAUUCAAAGGACUGUUUAUGUUUUUGUUCCGGUGUGACCGUAAUCCGAACAGGGACGCAGUCUGAUUUAA --------..............(((((.((((........)))).)))))(((((((....(((((((((((((.....)))......)))))))))))))))))....... ( -27.30, z-score = -1.79, R) >consensus ________UUAUAUUUAUUACUUUUGACUGCACAGUGGAAUGCAUUCAAAGGACUGUUUAUGUUUUUGUUCCGGUGUGACCGUAUUCCGAACAGGGACACAGUCUGAUUUAA ......................(((((.((((........)))).)))))(((((((....((((((((((.((.(((....))).)))))))))))))))))))....... (-25.29 = -25.30 + 0.01)


| Location | 15,697,880 – 15,697,992 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 89.63 |
| Shannon entropy | 0.13938 |
| G+C content | 0.38141 |
| Mean single sequence MFE | -23.53 |
| Consensus MFE | -18.36 |
| Energy contribution | -18.81 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.731845 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 15697880 112 - 22422827 UUAAAUUAGACUGUGUUCCUGUUCGGAAUACGGUCACACCGGAACAAAAACAUAAACAGACCUUUGAAUGCAUUCCACUGUGCAGUCAAAAGUAAUAAAUAUAAGCUCCUCA ........(((((((((((.....))))))))))).....(((................((.(((((.(((((......))))).))))).)).............)))... ( -28.85, z-score = -3.58, R) >droSim1.chrX_random 4146 104 - 5698898 UUAAAUCAGACUGUGUCCCUGUUCGGAAUAUGGUCACACCGGAACAAAAACAUAAACAGUCCCUUGAAUGCAUUCCACUGUGCAGUCAAAAGUAAUAAAUAUUG-------- ........((((((.....((((.......(((.....))).......))))...))))))..((((.(((((......))))).))))...............-------- ( -19.94, z-score = -0.95, R) >droSec1.super_37 278491 104 - 454039 UUAAAUCAGACUGCGUCCCUGUUCGGAUUACGGUCACACCGGAACAAAAACAUAAACAGUCCUUUGAAUGCAUUCCACUGUGCAGUCAAAAGUAAUAAAUAUAA-------- ........(((((.((((......))))..(((.....)))...............))))).(((((.(((((......))))).)))))..............-------- ( -21.80, z-score = -1.80, R) >consensus UUAAAUCAGACUGUGUCCCUGUUCGGAAUACGGUCACACCGGAACAAAAACAUAAACAGUCCUUUGAAUGCAUUCCACUGUGCAGUCAAAAGUAAUAAAUAUAA________ ........((((((.....((((.......(((.....))).......))))...))))))..((((.(((((......))))).))))....................... (-18.36 = -18.81 + 0.45)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:47:26 2011